Baby It's Cold Outside
Speaking of papers that need to find the light, I offer the following linked manuscript up to the science gods. This is a short paper that was orphaned from my dissertation, and this summer I challenged myself to write a manuscript on these data in one day. I actually took two, or parts of two, days knowing that it was only a tiny descriptive study of a limpet and perhaps no more interesting than the Genbank numbers contained within, but I guess I hope the data will be useful to somebody who is starting their graduate work and is considering work on Tectura testudinalis. David Reid is a wonderful colleague who I met at the CORONA meetings, and he was kind in his rejection, so I’m going to put this one into a web bottle and cast it to sea.
Wares_Tectura

The paper isn’t wrong, just insufficient. But the limpets are nice, so good luck little paper.
Our final news for the lab would be that we managed to earn three awards at the Genetics Department holiday party, I can’t remember which categories Christine’s entries were in but the vegan chickpea-spinach Indian-style dish I made got a blue ribbon in the vegetarian category, I’ll be sure to dogear that page in my cookbook!
Also if you are looking for entertaining reads on evolution and/or bees, check out the work of Jay Hosler (an image is on the “Invert Comics” page), a biologist who has combined his knowledge and skills as a cartoonist to generate some pretty interesting outreach.
Organization or the Lack Thereof
Today I’m coming to grips with the fact that a few hundred fairly important DNA isolations appear to have been lost somewhere in the last 3-4 years, they are some DNA samples from specimens collected in Chile in 2004. That is a bummer, because I may have an opportunity to get some next-gen sequencing done in the very very near future.... but I need some DNA! I have a technician starting in January to help organize these things, but it may be a busy next couple of weeks. Fortunately, I asked my students working on the first batch of Chile specimens to dissect away tissues and then keep the rest of the specimens in labeled vials. A real pain in the ass for them at the time, but a very much appreciated repository of information to me right now!
This has me thinking about how to organize samples in the future. Much of what we have is organized the way we would organize things when I was a grad student: it is in a lab notebook, or an Excel spreadsheet if we are lucky. That works fine for projects with samples in the low 100’s, but doesn’t work at all once the projects get larger, or have more loci, more locations, more times, and so on.
How complex do we want the process of curation to be? There are some really nice options, such as this LIMS plug-in for Geneious, which are worth considering as an all-in-one approach. However, the mental cost of getting started in that way is pretty high, both for me as advisor as well as for an incoming technician or graduate student. Most of the system only works well once you understand all steps of the process - and if you understand all steps of the process, you may also understand that Geneious doesn’t always handle each of these steps in the ideal way (though in general I’m a fan of that software).
For now I’m considering keeping things really basic - something like Excel or Numbers - which can then easily be uploaded into an actual database, either a baby one like Bento or a more complex one like that linked above. Either way it is clear it is time to get more savvy about what I’m freezing, because chances are good that any single freezer rack is containing a chapter for a graduate student, no reason for those hard-won samples to go to waste.
Chile and The Force

Three days in Chile, quite a weekend. I won’t say it was hectic, because I spent a lot of time waiting in airports and then a lot of time being well-cared-for by my hosts in Chile. We camped on the beach at Temblador, ate wonderful food, got to know each other well. My work in Chile dovetails very nicely with that of postdoc Jenna Shinen, who is currently based at ECIM in Las Cruces and works with Sergio Navarrete.

We are both studying the dynamics of the barnacles Notochthamalus and Jehlius, and this trip was an opportunity to get to know the system better (for me - Jenna knows it inside and out), meet the field technicians (Elliott and Felipe pictured below), and of course to collect some barnacles.


Along the way I had some fantastic seafood (mariscos), empañadas, good drinks, watched some fútbol.... and perhaps most surprisingly, got re-acquainted with Star Wars. These movies have gripped my Chilean colleagues pretty fiercely, and we spent a lot of time in the intertidal singing this:
It is amazing how easily one can fly around the world so quickly, stand in places that Darwin may have stood 150 years ago, study the same organisms....and have the most memorable component of the trip involve Darth Vader.
Wikipedia

Sure, it leads to all sorts of confusion and conflicts with students who browse Wikipedia for 10 seconds for the answer to a question (and worse, when they copy text from Wikipedia into their paper). Sure: it can be wrong. It is only as good as the people involved who are writing the text. It is not as clearly peer-reviewed as a journal article or a book, though it is most certainly “peer” reviewed. Bias exists everywhere in life, and as long as you are okay with the wabi-sabi nature of information as it filters out to us, Wikipedia is a really good place to start sometimes.
I’ll admit it, I’ve used it as the basis for some of my own knowledge as a biologist. I’ve also contributed to information that is out there. And frankly I’m completely okay with colleagues, students, and so forth using it in the same way I do: a starting point for discovery. As long as the little Bayesian in your head reminds you that the truth is probably only about 90% (in Wikipedia and in life) knowable, you can often figure out what you need to know, where you need to look to know it more certainly, and whether you are even searching under the right rock for this knowledge.
Anyway, maybe I’m just overtly justifying the fact that I’ve actually donated to the Wikipedia foundation, but I think it is an astounding bundle of admittedly imperfect knowledge about the planet that I’m pretty grateful for. How else do you satisfy the curiosity of a four-year-old (or, sometimes, a graduate student) if you don’t have this at your fingertips?
Need one more reason to like Wikipedia? Check out this page. I bet you’ll lose at least 10 minutes browsing there.
More AAPL Love
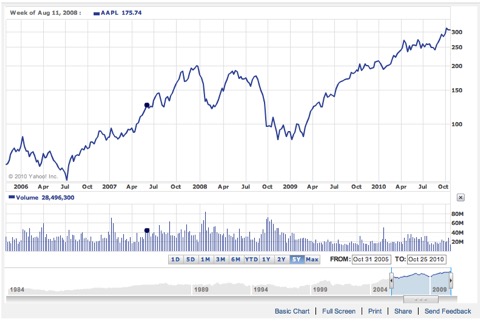
Above is the chart for Apple stocks over the time period that my lab has been operational. I’d like to think I have played some small part in that, since I’ve converted a few incoming students from the Windows world to Mac, bought a few big ones for the lab myself, and pitched the utility of having an iPhone while in Faculty Senate meetings.
More recently, to celebrate some good news (and to blow a little bit of summer salary), I bought an iPad. Yes, I’m riddled with guilt for buying a new gizmo, no matter how benign they claim the construction materials to be. I was worried that I was just going to find it as another toy, and assumed I could always sell it if it wasn’t truly being useful.
I have to say now, having had it for a month, it is actually a really nice part of my “productivity”. Administrators, please don’t laugh! I’ve been effectively paperless since getting it, reading and marking up a couple dozen PDFs, reading books (and most recently a monograph on the biology and oceanography of the Humboldt Current System), and even reading Science updates on there. I can take decent notes with the Note Taker HD app, and I’m learning how to use tools like 2Screens to give presentations - I think this will be useful for teaching, as I can annotate my cryptic slides on coalescent theory in real-time.
Of course, I could have done that with an overhead projector, but I don’t know if we even have one of those anymore....
Anyway, after getting over my bashfulness about bringing it to lab group and seminar, I find it nicely indispensable. It is more friendly to have in meetings than a laptop that you hide behind, though of course it has its limits. Then again, a laptop isn’t as easily handed to a 4-year-old as an iPad is, with nicely intuitive games and puzzles that are actually decent educational material. I wonder if any of our sequence alignment software will get ported to the iPad soon....?
Technology
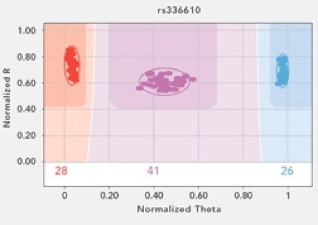
This week has been a good one in the Wares Lab, not least of which because a huge financial gamble has paid off, and is a great indicator of how population genetics and phylogeography is changing. I received some funding from the University of Georgia Research Foundation, intended for junior faculty to generate preliminary data for bigger fish (NSF, NIH) proposals. The entire goal of the award was to try out the Illumina Golden Gate genotyping technology with the Streblospio transcriptome that Christina Zakas is working with. We nervously sorted and re-checked the SNPs in the data, and weeded it out to a set of about 200 that we hoped would be applicable ... and more importantly, scorable. This species has been really recalcitrant when it comes to successful PCR. Fortunately, the Illumina technology doesn’t really go through PCR to get its genotype! In the end, 96 loci were chosen and the array was developed (again, at a big cost for a technology that we’ve never worked with or even seen worked with around here, on a species that we couldn’t even reliably amplify mitochondrial genes).
The data started rolling in this week, and it is phenomenal. Of course some of the loci don’t work well - but around 80% do, which is a better fraction than I’ve ever had when developing primers for nuclear loci in these non-model critters previously. And some DNA templates are not great quality, but close to 95% appear to be just fine. This puts us in position to suddenly be studying these tiny squishy mud worms at a global scale with 2 orders of magnitude more data than ever before. Money well spent, and we’ll be using the same technology on upcoming projects.
Marine Biology
McSweeney’s knows it is funny.
Seinfeld knew it was funny.
And with colleagues like J-Long, it IS funny.
After the hairy theory/numerical/simulation Genetics paper our lab group read today though, I’d rather call myself a marine biologist any day!
North Atlantic

The above image was snared from the website of Geoff Trussell’s lab at Northeastern University. Geoff is one of my many great colleagues that I know from interactions at the CORONA meetings and every year that I can make it to the Benthic meetings on the east coast (and really, who would want to miss these meetings? Nice one, Jeremy...). Anyway, the image is primarily of the dogwhelk, Nucella lapillus. A really beautiful snail, lots of color variation, interesting genetic patterns, part of my dissertation.
The work is finally being updated, with modern statistical (ABC) approaches, by my good friend and colleague Mike Hickerson and his student and postdoc (particularly lead author Katriina Ilves). The results are clearly starting to change. There are taxonomic controls on who survived glaciation and who didn’t - on the east coast of North America, at least - rather than larval life history controls, if the statistics tell the whole story. What is amazing is how big of an impact that paper had in 2001, and I suspect this paper will have a big impact in 2010.... and yet we still clearly know so little. Most of these species have still only been assayed for mitochondrial variation. The time will come soon when we can apply for funding to tackle the same community using variation across the entire genome.
Meetings Meetings
One trade-off involved with working out such details is that in a good collaboration, the workload is appropriately shared so that the best possible science is done most efficiently. In this case, my standing collaboration with Sergio Navarrete has led to a near-perfect situation where his postdoc happens to already be sampling many of the sites that we need samples from in the coming years. This is all going to work out well.... except now I have fewer good excuses for research jaunts to coastal Chile! Clearly, some of our face-to-face meetings will need to be in person, perhaps over a good plate of local seafood and some pisco sours.
Communication Breakdown
While the title of this post stems from Led Zeppelin (but could also be a catchy bluegrass title), it is clear that communication is one of the most important components of science. There are inestimable differences in what can be accomplished in a simple, face-to-face conversation compared to email or even a phone call. So, even though I complain about the travel (see my July 7 post), I know there is good to be had (and not just the beer drinking) in going to meetings.
Some meetings are fun, and a lot of ideas are exchanged – but it never quite translates to getting things done. I’ve gone to a lot of “working group” type meetings, 20-30 people, and it is hard to convey the speed with which ideas loft on hot air (and can be quickly shot down) at such sessions. Sometimes these groups end in synthesis papers, white papers, and such, but it is too easy for us to lose focus once we return to our desks and our full inboxes.
Last week, upon learning that my Chilean colleague Sergio Navarrete was visiting the University of South Carolina, I buckled up and drove 6-7 hours round trip so I could have lunch with him and visit with some of my favorite U.S.-based marine colleagues for a bit as well. Nearly a full day of driving is usually anathema to me, a waste of fuel and hard on my back. But it was extremely productive, even though we spent 2/3 of our time talking about the earthquake that struck Chile in February we sketched out some great ideas for how the field component of our work can begin soon.
I was lucky there - having a necessary and enjoyable colleague within driving distance, and a day when I wasn’t teaching, doesn’t come together often. So now I’m struggling with how to manage my collaborations across the globe most effectively. Our EID work is underway, and we’ve had two meetings using Adobe Connect Pro (to connect the Athens folks with Katie Patterson Sutherland at Rollins College; eventually we’ll pull in more partners to these meetings too). The first meeting went well, and I was impressed with the software. Yesterday - not so well, some echo effects and hiccups in attempting to connect more than 2 laptops, made me a little more concerned.
I think this is going to be fairly important for my OCE funded work in Chile. Two of us are here at UGA, one colleague in New Hampshire, others in Chile. Teleconferencing (either the institutional route, or up to 5 lines on my cell phone) has strange “who has the conch?” problems (and costs associated with long-distance calling; iChat only works on Macs; Skype only allows 2 video feeds. I briefly tinkered with Cisco’s WebEx, which works similarly to Connect Pro, but it seems we are likely to stick with the latter for the EID work so I’d rather know more about one imperfect system than just enough to be dangerous about two such systems.
Hopefully these Voice/Video Over IP solutions are going to be improving. It’s certainly going to be key for us getting things done in the coming years.
update: Ron Eytan, new postdoc in the Wares and Lipp labs, pointed me toward what appears to be an excellent solution. EVO is “worldwide collaboration network” from Cal Tech. I’m not sure how they do it, but the interface is easier, the video is much higher quality than the above solutions, and the audio is no worse....and it is FREE!
Up-and-comers

The image above is from Iliana Baums’ lab - one of several working on the imperiled elkhorn coral, Acropora palmata (and really, most of the rest of us are following the lead of Iliana and her students!). This picture was taken in Puerto Rico, and I snagged it from the web to commemorate another addition to the fleet of researchers working on these corals. SUNFIG student Jesyka Meléndez, who spent the summer of 2009 in my lab learning molecular techniques and exploring a metapopulation (or at least thats what we’re calling it for now) of Melampus bidentatus on the Georgia coast, is from the University of Puerto Rico - Cayey. She did a great job in my lab as a freshman, and has now been awarded an EPA fellowship to study pollution and sedimentation on Acropora reefs in Puerto Rico. It’s actually way more fun to see what young enthusiastic people can do, and want to do, than to track my aging colleagues and their publications.
For that matter, here is a 16 year old who has done a high school research project that is essentially our big EID project on a severe budget. It blows my mind what students have access to now, compared to the things we were stuck with when I was in high school (and I’m not that old). You can do comparative (or human) anatomy on an iPad now, you can easily do PCR and electrophoresis in a high school lab, basically kids have the opportunity to do and see anything now. It blows my mind, especially with imagining what my almost-4-year-old will be doing in the next 20 years!
Famous Colleagues
Then yesterday I learned about fellow Duke grad student (and fellow UC Davis postdoc, and fellow fish biologist, invasive species biologist, salt marsh biologist - what doesn’t he do?) Mike Blum appearing on the Colbert Report. Mike’s interview, about the toxic effects of the oil spill and dispersants used for months afterwards, starts at about 11:08 but it is worth tuning in at about 10:00 for the jokes about “shrimp taint”.
Epistemology
We (the Genetics department, more specifically Brian Condie and myself) are teaching a new course this year for our incoming graduate students that will be playing around with this theme as well. It is called “Problems in Genetics” but the goal is not to learn the latest in quantitave PCR approaches, but to learn more about how good questions are asked, and why. We’ve started the semester off with a chapter from E.O. Wilson’s Consilience, and it may have been a big surprise to the students to be reading something like this in a Genetics class but I hope this pays off. We really want to break our new students of some of the habits that undergrad scientists get into. They need to become skeptical of the literature while understanding its value; they need to understand how to develop questions themselves; and they need to understand the conceptual breadth of our field, and how it attaches to other fields like ecology and biochemistry.
We’ll be revisiting old experiments and how they guide us today; discussing what models are, and the value of both conceptual and mathematical approaches. Most of all, we want to finally squash the lack of understanding that affects almost every academic department, the narrowness that leads to disdain for compatriots simply because they aren’t studying something so directly tied to what we are doing. Put simply, we want a student to understand why somebody is asking the question and using the methods, even if that student doesn’t care to ask that question or use that method themselves.
Wish us luck. Welcome to the new school year.
More iPhone Love
Portland
The only conundrum that remains from great meetings like this - aside from how to respond to all the great ideas that come from talking to so many talented colleagues - is the ugly environmental side of meetings. The organizers in Portland went to lengths to put us somewhere that required no driving, they encouraged bringing reusable cups, and Portland is an extremely green city. But the carbon footprint of flight cannot be ignored. There is no replacement for face-to-face interactions at these meetings; in particular, I’m thinking of some of the extraordinary grad students I talked with over the course of three days about their work and their future plans. But, we can’t ignore the impact of air travel, which sadly negates many of the things we struggle towards with our reusable cups, cycling to work, and unplugging unused appliances. The figure below was taken from a recent story in Mother Jones, and makes me queasy about the 2 flights I’ve already committed to later in 2010, and probably another 1-2 in the next year.
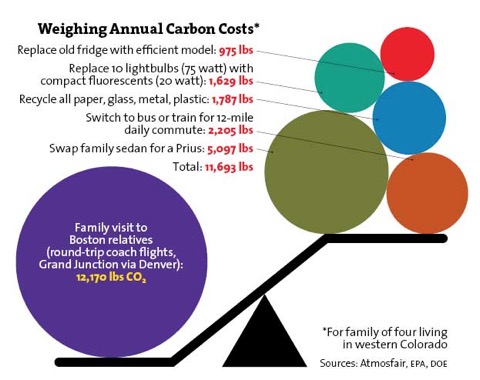
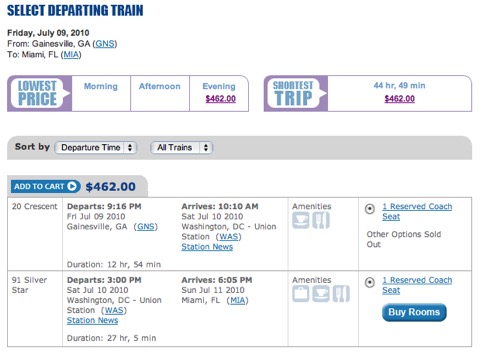
What I am going to do is something that is good for me and my family as well as for the environment. I don’t like being away too much - sometimes it is necessary, but sometimes as scientists we accept invitations to do things that mean little for our work (and more for our egos or careers). If Steve X invites me to give a talk at Fantastic University, it may be flattering and I may gain professionally, but it is not generally critical for my life or my ongoing projects. So, I’m capping myself to 4 air trips a year. That may be tough. I may have to drive to some places I ordinarily would fly to, or figure out Amtrak in the southeastern U.S. (it isn’t good, we are far behind Europe and California in this regard, look at this schedule and price for a trip to Miami!).
I hope my colleagues understand, but having this excuse is also good for making sure I’m home with my family just a little bit more as well.
Thank You Apple
Be Careful What You Wish For
Suffice it to say, I’ll be looking for help. Maybe a technician, maybe another grad student. Maybe somebody to mow my lawn, since I’m not going to have as much time around the house for a few years it seems (one can wish; it is supposed to be 99° today!)...
beautiful nauplius drawing from Perez-Losada et al. 2009 BMC Evol. Biol.
Dirty Water
Obviously I’m talking about the environmental catastrophe that British Petroleum allowed to happen. I don’t think I’m the only one of the opinion that not enough safeguards were in place given the known and documented risks of the well that is currently dumping more than a half-million gallons of oil into the Gulf of Mexico every day, for more than a month now. The spill is ruining the marshes of the Gulf Coast (a disaster for so many people, including scientists studying these habitats), and may now be encroaching on the Florida Keys. There are other UGA marine scientists studying where the oil is going (link to Mandy Joye’s blog, and now interview on NPR), and so we are starting to get a clearer picture of just how broad are the effects of the oil spill and subsequent use of hazardous dispersants.

What will this mean for our own project on the microbes living in the Keys? Will the white pox epidemic even matter? These are questions that I guess we are poised to figure out. Hopefully we can get some of our sampling done soon down there, and then use available resources (including research funding being released from BP?) to sample again after the oil and dispersants have washed through. We all need to be documenting what is going on so this information can be used to prevent it from happening again.
It all makes me sick.
Startup
I have a Bio-Rad vertical gel rig that, to my knowledge, hasn’t been successfully incorporated into a research project in 5 years. That is close to $5,000 worth of a pain-in-the-neck gizmo that just doesn’t perform half as well as the vertical rigs I used in Tom Turner’s lab, combined with the shift in methodology that has put SSCP and other fragment analysis protocols way back on our back burner. But I can’t sell it on eBay....here it shall sit, hopefully we can find a use at some point. It has a nice big buffer tank, perhaps I should catch some minnows and make a small aquarium out of it (with temperature control!)?
How many frantic purchases with Applied Biosystems or other companies did I make before realizing that each one of those purchases had a hidden $30-40 shipping cost (dry ice, overnight)?
What about the half-broken chair I inherited in my office? I was told that furniture purchases on my startup funding HAD to be through Chastain’s, a furniture place in downtown Athens. Nothing against that business, but the amount of time I spent bouncing on the 5-6 models available in their showroom, only to leave with a $500 chair charged against my startup, would have been better spend buying three chairs from IKEA on my own credit card. Then I could have had different colors for my changing moods as a young professor.

Most importantly, I would have loved to have had the same amount of money with so few strings attached NOW. Just the few examples of poor expenditures noted above, add those up: I could have a draft transcriptome of any species I want for that amount of money. I could have picked a small group of related species, say the freshwater mussels that we worked on, and dramatically improved our marker availability, understanding of molecular evolution, and so on for just 1/10th of that initial startup package.
But that’s how science rolls. We make mistakes. Technology changes. And we keep writing grant proposals to move on.
New Site
It has been a week of banging my head against computational things in lots of ways. Turns out, for example, that MrBayes does not fully compile correctly under 64-bit architectures (like Snow Leopard OS 10.6 on my Mac Pro), so I lost a few days of work on freshwater mussel phylogenies figuring that issue out, and ultimately just snagging the compiled version off of my old G5 (32-bit) and everything works fine now, thanks. Maybe someday we’ll actually get these mussel papers out, I’d like to do it soon so I can keep working on them. Fun excuse to go splashing in a river in the summer!
Worms

A little proof that I do escape from behind my monitor from time to time. Two weeks ago, I was out at Sapelo Island to visit our Georgia Coastal Ecosystems sites in hopes of finding Streblospio benedicti for my student (Christina Zakas). My field assistant mostly liked to help with the hose and mud. The procedure for finding these critters gives lie to the phrase “find ‘em and grind ‘em” as applied to phylogeography. The problem is that people have come to assume such studies are as simple as driving to an area, scooping up an organism, and then squeezing DNA out of it. Truth is - and its good to be reminded myself when I’m reviewing papers for journals - small sample sizes often result from organisms that are very hard to find, have nasty associated chemistry that inhibits PCR, and/or organisms that are threatened (and thus rare, though we want the data ever so much more).
A week later, I got to touch the Pacific as well. I just got back from an NSF workshop on “Evolution and Climate Change in the Oceans”. Phenomenal group of scientists, very high-level discussion and ideas, a lot of fun - and a good excuse to spend a few days on Santa Catalina Island, off the coast from Los Angeles. My only regret is forgetting to bring running shoes, after several days of travel and meetings I feel pretty sluggish.
Back in the Saddle
Also, it seems I’m becoming more permanent around here, as my collaborations have become official: the Bio-Oce proposal with Jeb Byers and Jamie Pringle is funded and starting in June, and my collaboration with Jim Porter and Andrew Park (Ecology), Erin Lipp (EHS), and Katie Sutherland (Rollins College) was also just tabbed for funding on April 1 (we think it isn’t an April Fool’s joke....)!
So, for all the high-tech wizardry I’ve proposed in the past, ultimately I’m thrilled to be funded essentially to do biogeography and community ecology. That is what got me started down the academic path in the first place, really. Stay tuned!
Busy February

But today I actually wanted to give props to my Ph.D. advisor, I left his lab 10 years ago now. It has been a good week or two for Cliff Cunningham, with notable mentions in Science for the collaborative work he has done with Scott France and others on the origins of deep sea corals, and now in Discover magazine where his work to help determine the true phylogeny and ancestral relationships of the arthropods has made big waves! The picture of the xenocarid above represents the closest living relative to the Hexapoda, including insects - so we’re talking about a deep relationship here, appropriate since these blind creatures are found in caves....
The First!
Honors Students

Oh, the people we deal with in this line of work. There are days when it can drive you nuts - politics, broken temp-control rooms, paperwork - but the students keep me on my toes in a lot of good ways. For New Year’s Eve I got to catch up with one of my former Honors students, John Binford. Currently a med student at Yale, budding proprietor of a pedicab business and a microbrewery, taking sanctioned sabbaticals to the Dominican Republic to work on nutrition improvement projects with the people. It is always amazing to soak in the energy that our best students bring to the table. Another of my former Honors undergrads is working on her Ph.D. now in entomology, and again I think I gained from the enthusiasm that she brought to her work in the field, in the lab, and getting ready for riding across the country on her bike (actually, John was on that trip too; and another undergrad that I worked closely with in her senior year also rode across the country....with the legacy of Fred Birchmore at UGa, maybe I should be planning my own post-tenure-decision trip....). Anyway, here’s to the beginning of another semester, and I hope I can bring back as much energy as I can from the holidays!