freezer
Still Have Mojo
26/02/11 14:13
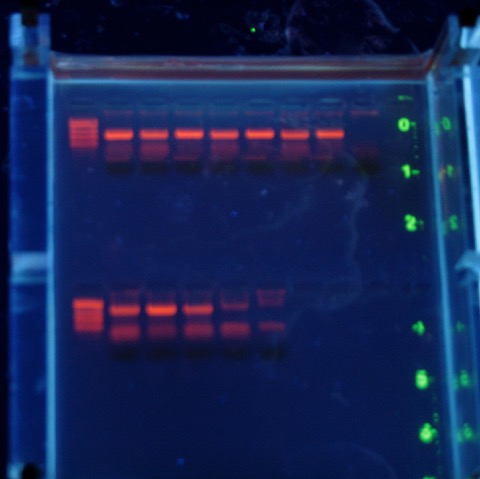
I love teaching the Evolution lab class at Georgia. I really do. But this class has led to more sleepless nights over the past 6 years than any other; why? Because maintaining a good flow of information, education, and fun in a lab class is tricky. And while failure is a large part of science, it doesn’t play well when you have a limited number of opportunities to teach and a number of topics you want to teach.
This year GENE 4230 will be repeating Peter Marko’s 2004 work on red snappers. This paper showed that much of what we buy at the store may actually be Lutjanus campechinus; but much of it is not. Fish fillets sold at red snapper prices are sometimes other (rarer) snapper species. Sometimes they aren’t snapper at all, but less valuable fishes. And all of that ambiguity leads questions about how best to manage this threatened stock. Currently, the fishery is closed on our Atlantic coast.
The problem? When I had Kelly double-check my protocol a couple of weeks ago to make sure the PCR would work, it did not. We tried a couple of different protocols, no dice. Finally I decided I needed to jump in and check some things. Long story short? Both our original primer stock, and the last batch of red snapper DNA we had used for this lab 2-3 years ago, were heavily degraded. They had been kept in the aging freezer in the teaching lab, so Genetics department head: there is something that we need to buy this year!
I’m pleased that I could quickly figure out what went wrong, and from the gel above it looks like the lab should work just fine (in case you are wondering, those are the short ‘universal’ fragment primers on top; the CB-12 and -13 primers from Marko’s paper below). There is a little smeariness in the negative control but I also didn’t change my tips in my rush to get this done today (Saturday....and it is 75° outside and sunny). No problem with the top set of reactions, that means I’ll be sleeping much better tonight.
Organization or the Lack Thereof
30/11/10 12:49
In five years I’ve managed to fill my $15,000 ultracold freezer, empty bits out of it, fill it again. There are 2 other freezers in our equipment hall, and one in the main lab. That is a lot of specimens, most of which are only locally organized - that is, they are organized in the brains of the people who put them in there, but not in any global sense. Sometimes, people don’t label things well, and sometimes things get lost.
Today I’m coming to grips with the fact that a few hundred fairly important DNA isolations appear to have been lost somewhere in the last 3-4 years, they are some DNA samples from specimens collected in Chile in 2004. That is a bummer, because I may have an opportunity to get some next-gen sequencing done in the very very near future.... but I need some DNA! I have a technician starting in January to help organize these things, but it may be a busy next couple of weeks. Fortunately, I asked my students working on the first batch of Chile specimens to dissect away tissues and then keep the rest of the specimens in labeled vials. A real pain in the ass for them at the time, but a very much appreciated repository of information to me right now!
This has me thinking about how to organize samples in the future. Much of what we have is organized the way we would organize things when I was a grad student: it is in a lab notebook, or an Excel spreadsheet if we are lucky. That works fine for projects with samples in the low 100’s, but doesn’t work at all once the projects get larger, or have more loci, more locations, more times, and so on.
How complex do we want the process of curation to be? There are some really nice options, such as this LIMS plug-in for Geneious, which are worth considering as an all-in-one approach. However, the mental cost of getting started in that way is pretty high, both for me as advisor as well as for an incoming technician or graduate student. Most of the system only works well once you understand all steps of the process - and if you understand all steps of the process, you may also understand that Geneious doesn’t always handle each of these steps in the ideal way (though in general I’m a fan of that software).
For now I’m considering keeping things really basic - something like Excel or Numbers - which can then easily be uploaded into an actual database, either a baby one like Bento or a more complex one like that linked above. Either way it is clear it is time to get more savvy about what I’m freezing, because chances are good that any single freezer rack is containing a chapter for a graduate student, no reason for those hard-won samples to go to waste.
Today I’m coming to grips with the fact that a few hundred fairly important DNA isolations appear to have been lost somewhere in the last 3-4 years, they are some DNA samples from specimens collected in Chile in 2004. That is a bummer, because I may have an opportunity to get some next-gen sequencing done in the very very near future.... but I need some DNA! I have a technician starting in January to help organize these things, but it may be a busy next couple of weeks. Fortunately, I asked my students working on the first batch of Chile specimens to dissect away tissues and then keep the rest of the specimens in labeled vials. A real pain in the ass for them at the time, but a very much appreciated repository of information to me right now!
This has me thinking about how to organize samples in the future. Much of what we have is organized the way we would organize things when I was a grad student: it is in a lab notebook, or an Excel spreadsheet if we are lucky. That works fine for projects with samples in the low 100’s, but doesn’t work at all once the projects get larger, or have more loci, more locations, more times, and so on.
How complex do we want the process of curation to be? There are some really nice options, such as this LIMS plug-in for Geneious, which are worth considering as an all-in-one approach. However, the mental cost of getting started in that way is pretty high, both for me as advisor as well as for an incoming technician or graduate student. Most of the system only works well once you understand all steps of the process - and if you understand all steps of the process, you may also understand that Geneious doesn’t always handle each of these steps in the ideal way (though in general I’m a fan of that software).
For now I’m considering keeping things really basic - something like Excel or Numbers - which can then easily be uploaded into an actual database, either a baby one like Bento or a more complex one like that linked above. Either way it is clear it is time to get more savvy about what I’m freezing, because chances are good that any single freezer rack is containing a chapter for a graduate student, no reason for those hard-won samples to go to waste.