2011
Fecundity 4
18/11/11 09:30

That’s Dr. Zakas next to me, successfully defended - in fact, she did such a good job on her dissertation I think the committee would have preferred to skip the defense and just head for the pub. Congratulations Christina!
Christina is going to be helping me in the coming months wrap up some analyses on the RAD-tagged data for Notochthamalus. I have to say right now I’m a little disappointed with the whole Nex-gen world, for the third time in as many attempts at getting 454 or Illumina data a huge portion of the data came back as junk (this time apparently a bubble in the line). I guess it will take a while for all the kinks to get worked out of these systems, and by then they’ll have invented something new!
Teaching
30/09/11 13:09
I’ve just about finished my first stand as lecturer in our Intro Evolution course at UGA, a class that started with 197 students... I think it is safe to say there are fewer now after the first exam. I have one lecture to give and am really grateful to the students as they put up with me figuring out the dynamics of a class that size, I’m only now getting more comfortable with singling people out so I get answers to my questions that are more audible than a group mumble. Yesterday I said, “You, Duke shirt” to get a response from a woman wearing a Duke football jersey. I mean, if you don’t want to get called on in class you have to wear something more inconspicuous than the football jersey for a school that barely plays football :)
Our labs genome data are slowly coming in; all of the Illumina data for Serratia are back, and Ron Eytan is busily figuring out how to assemble the bacterial genomes. The Georgia Genomics Facility is having some technical difficulties with the 454 sequencer, so that project has been slowed down considerably. And hopefully soon the data will be back for the work in Chile, which - now that I think about it, I leave for Chile in less than 3 weeks, so having those data would be nice!
Our labs genome data are slowly coming in; all of the Illumina data for Serratia are back, and Ron Eytan is busily figuring out how to assemble the bacterial genomes. The Georgia Genomics Facility is having some technical difficulties with the 454 sequencer, so that project has been slowed down considerably. And hopefully soon the data will be back for the work in Chile, which - now that I think about it, I leave for Chile in less than 3 weeks, so having those data would be nice!
Leaving!
29/08/11 10:58
Well, today ended up being the day that John Robinson and Ron Eytan left the lab, both going on to new postdoctoral experiences. I know we are going to get a lot of papers out together, and I will definitely miss the regular exchanges of ideas (and occasionally bourbon) with these two!
John is heading on to a postdoc with Greg Moyer at the USFW facility in Warm Springs, GA.... as best as I understand this will be meta-analytical work on genetic patterns in the southeast, something that John - with his statistical and computational abilities in R and other environments - is very well suited to. Ron is going to continue working on our Serratia genomes while up at Yale working with Tom Near on big fish phylogenies - lots of fish, lots of genes, tons of data, again something ideal for his talents and interests. Good luck to both of you, and hope to see some manuscripts from you soon :)
John is heading on to a postdoc with Greg Moyer at the USFW facility in Warm Springs, GA.... as best as I understand this will be meta-analytical work on genetic patterns in the southeast, something that John - with his statistical and computational abilities in R and other environments - is very well suited to. Ron is going to continue working on our Serratia genomes while up at Yale working with Tom Near on big fish phylogenies - lots of fish, lots of genes, tons of data, again something ideal for his talents and interests. Good luck to both of you, and hope to see some manuscripts from you soon :)
All your base are belong to us
15/08/11 10:00
It occurred to me that we are very close to having whole genomes for 48 microbial strains, and 50x coverage for a different set of 48 microbial samples from coral reefs in the Keys, and 50x coverage across each of 16 populations of RAD-tagged barnacles from Chile. By the end of the year. Something like 200,000,000,000 nucleotides that we have to analyze and make sense of in the coming year. If each one was a penny, stacked on top of each other it would just about reach to the moon. Good thing they aren’t that big.
Art
30/06/11 14:24
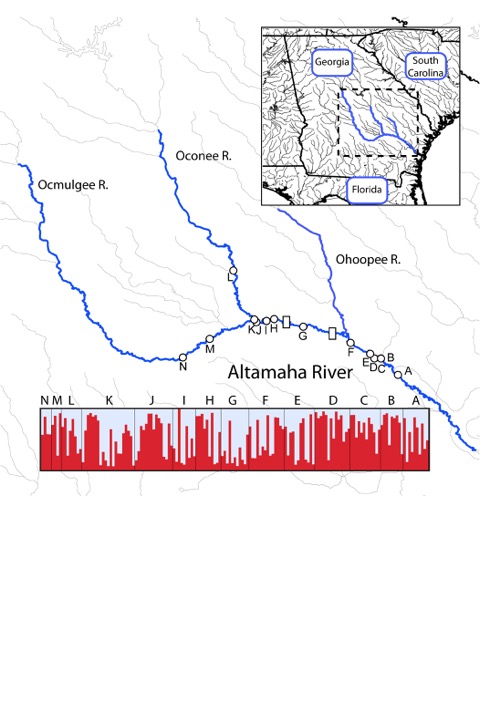
I may be lousy with a brush or pencil, but I do like making figures. Here is one I made today that uses R to plot data from the Scenic and Wild Rivers maps with state boundaries, along with the lat-longs of sample locations for the freshwater mussel Elliptio dariensis, and a Structure genotype plot to match the locations (prettied up with Adobe Illustrator):

Stats
27/06/11 11:47
Summer is here so there isn’t a ton to report. I’m the new Graduate Coordinator, so have to think of new ways to bug the students! One thing that we try to bug our students about is their statistics background. I promise you, whoever you are reading this, that if you are still in the formative years of your career - or even if you are old like me - you’ll probably be better off if you understand statistics and probabilities a little bit better. If nothing else, you’ll be able to interpret political polls better, and will do better when you visit Vegas. But in the meantime, old friend and colleague Alisha Holloway recently posted this from a bioinformatician’s blog, and it summarizes what I’m talking about better than I can....

(xkcd, of course)
Oh, one bit of newsworthy lab-ness. We finished the BeadXPress run with the 5th plate about 6 months past its “expiration date”. We were nervous as to whether this date was just CYA by Illumina or if the reagents really would decay, but it turns out that our genotype rate was just fine on the last plate so Christina is set to finish her dissertation soon!
Also, I don’t know why the style is messed up on my website. Will fix.... someday.

(xkcd, of course)
Oh, one bit of newsworthy lab-ness. We finished the BeadXPress run with the 5th plate about 6 months past its “expiration date”. We were nervous as to whether this date was just CYA by Illumina or if the reagents really would decay, but it turns out that our genotype rate was just fine on the last plate so Christina is set to finish her dissertation soon!
Also, I don’t know why the style is messed up on my website. Will fix.... someday.
Gloves
13/05/11 14:09
Well, we are having sequencing problems again. Very frustrating. The outcome looked a lot like the problems we were having earlier this year, using old (about a year old) BigDyes. I bought more in early March, and everything went great for a while.
Then the past 2-3 plates were bad again. Kelly and I did some sleuthing today and traced all the problems back to any sequencing that happened after April 27, 2011. On that day I sent a message out to the lab noticing that the autoclave gloves had gotten stuck in the seal at the top of the freezer (very close to where our pricey reagents live) and the freezer was thawing. Notice that even today the gloves are in the same place..... OK, I just moved them.

Anyway, I think our beloved BigDye must have sat at an elevated (thawing) temperature for 12 or more hours, and things haven’t been so good since. I hope it fixes things to buy a new $800 tube of reagent, because otherwise this constant problem-solving process is making my brain hurt.
A couple of links of interest: my colleague Sarah Gilman (of former Grosberg lab fame) has a nice paper out on the predictability of organisms shifting their range in response to climate change (other friends/colleagues like Leslie Rissler are in there too), here is a page pointing to that paper. And for anybody who likes to think about the response of the planet to past glaciation, the recent earthquakes in Maine are a nice reminder that nothing is static, it is always in flux.
Then the past 2-3 plates were bad again. Kelly and I did some sleuthing today and traced all the problems back to any sequencing that happened after April 27, 2011. On that day I sent a message out to the lab noticing that the autoclave gloves had gotten stuck in the seal at the top of the freezer (very close to where our pricey reagents live) and the freezer was thawing. Notice that even today the gloves are in the same place..... OK, I just moved them.

Anyway, I think our beloved BigDye must have sat at an elevated (thawing) temperature for 12 or more hours, and things haven’t been so good since. I hope it fixes things to buy a new $800 tube of reagent, because otherwise this constant problem-solving process is making my brain hurt.
A couple of links of interest: my colleague Sarah Gilman (of former Grosberg lab fame) has a nice paper out on the predictability of organisms shifting their range in response to climate change (other friends/colleagues like Leslie Rissler are in there too), here is a page pointing to that paper. And for anybody who likes to think about the response of the planet to past glaciation, the recent earthquakes in Maine are a nice reminder that nothing is static, it is always in flux.
Outreach 2
03/05/11 11:38
Wow. Today I went to Fowler Drive Elementary and met with the 4th graders, after being invited by teacher Frannie Gay. I brought some slides to show on my iPad (today’s fourth-grade classrooms are SO cool compared to the dreary situation I went through 30 years ago; they have integrated LCD projectors with a projection screen that doubles as touch-pad) and the kids must have asked me 100 questions about corals, sharks, bacteria, washing hands, Finding Nemo, and dirty water. Mission accomplished.

Faculty from UGA do this each spring as part of our “LemonAid” program, started a couple years ago when we were being furloughed thanks to lousy budgets. When life gives you lemons, get it? So on furlough days faculty would go to local schools; this year, no furloughs but it seems a good tradition to keep up. And, it gives me one of those extremely rare opportunities to not just claim to be a marine biologist, but actually feel like I’m doing something that might change something for the better on this world. Maybe one of those kids will get the overall message about clean water and take it to heart.

Faculty from UGA do this each spring as part of our “LemonAid” program, started a couple years ago when we were being furloughed thanks to lousy budgets. When life gives you lemons, get it? So on furlough days faculty would go to local schools; this year, no furloughs but it seems a good tradition to keep up. And, it gives me one of those extremely rare opportunities to not just claim to be a marine biologist, but actually feel like I’m doing something that might change something for the better on this world. Maybe one of those kids will get the overall message about clean water and take it to heart.
Outreach
11/04/11 10:17
Although in principle it should be easy to get people excited about science - tracking pathogens through an endangered coral population, monitoring genetic diversity to understand the larval dispersal of species on the Chilean coast - it is really tough to translate this excitement to the general public. XKCD seems to get it.


Fecundity 3
01/04/11 11:38

Congratulations to Dr. John Robinson, who just finished the easiest Ph.D. defense I’ve yet seen. John came to my lab just less than 5 years ago, and has been a phenomenal addition to our little population here on the third floor and the department as a whole. I won’t, however, miss feeding his Daphnia lines.
This is just a short note between meetings, as we have our final spring faculty meeting in about 15 minutes. I’ve also come to the sad conclusion that BigDyes sequencing reagents do in fact age in the freezer. As my old funding wrapped up and my new funded projects began, I had two full tubes of BigDyes in the freezer, enough to keep us going for a few months - and some new students working on sequencing projects. As the data came back and weren’t very pretty, I thought it might be novice errors, but this week has finally proven that it is time to toss $1800 worth of reagents into the trash.
Flying Barnacles
04/03/11 13:42
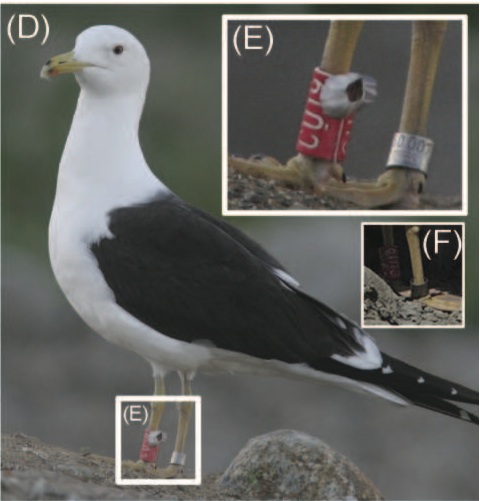
from Tøttrup et al 2010 Biofouling 26: 577-582. Thanks to Christine for finding this...although the individual in (E) looks dead, some of the other images in the figures in this paper are more likely alive and living a life most barnacles only dream of.
Still Have Mojo
26/02/11 14:13
I love teaching the Evolution lab class at Georgia. I really do. But this class has led to more sleepless nights over the past 6 years than any other; why? Because maintaining a good flow of information, education, and fun in a lab class is tricky. And while failure is a large part of science, it doesn’t play well when you have a limited number of opportunities to teach and a number of topics you want to teach.
This year GENE 4230 will be repeating Peter Marko’s 2004 work on red snappers. This paper showed that much of what we buy at the store may actually be Lutjanus campechinus; but much of it is not. Fish fillets sold at red snapper prices are sometimes other (rarer) snapper species. Sometimes they aren’t snapper at all, but less valuable fishes. And all of that ambiguity leads questions about how best to manage this threatened stock. Currently, the fishery is closed on our Atlantic coast.
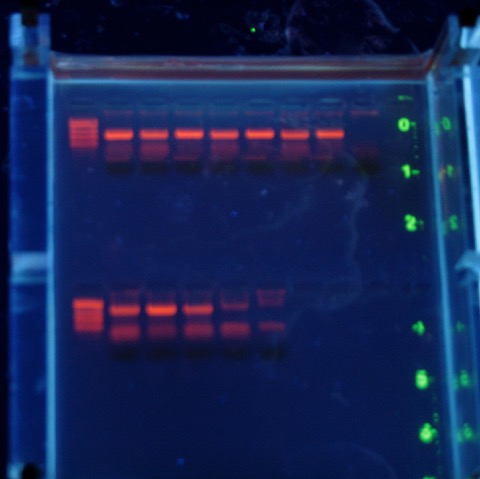
The problem? When I had Kelly double-check my protocol a couple of weeks ago to make sure the PCR would work, it did not. We tried a couple of different protocols, no dice. Finally I decided I needed to jump in and check some things. Long story short? Both our original primer stock, and the last batch of red snapper DNA we had used for this lab 2-3 years ago, were heavily degraded. They had been kept in the aging freezer in the teaching lab, so Genetics department head: there is something that we need to buy this year!
I’m pleased that I could quickly figure out what went wrong, and from the gel above it looks like the lab should work just fine (in case you are wondering, those are the short ‘universal’ fragment primers on top; the CB-12 and -13 primers from Marko’s paper below). There is a little smeariness in the negative control but I also didn’t change my tips in my rush to get this done today (Saturday....and it is 75° outside and sunny). No problem with the top set of reactions, that means I’ll be sleeping much better tonight.
Barnacle Mating
21/02/11 09:55
One of the great things about going to big meetings always used to be, frankly, partying with my old comrades from grad school. As we age, I have to say I am starting to get just as excited about interacting with the up-and-comers who are transforming science as we watch. Though this is a belated shout-out to her, I met UC Davis graduate student Morgan Kelly at those meetings and was really impressed with her work on Tigriopus. I was even more surprised last week when one of my newer grads brought this paper to me. I’ve spent most of my research on acorn barnacles and thought I knew them pretty well, but this paper really opens up the door toward understanding reproductive behavior and strategies in this diverse group. What is particularly intriguing is thinking about some systems I know about where parasites can induce a change in reproductive strategy, e.g. from hermaphroditism to androdioecy. Whether this turns into a new project or not it is too early to say, but thanks to Morgan and Eric Sanford for a really interesting read!

Answers Lead to Questions
10/02/11 09:30
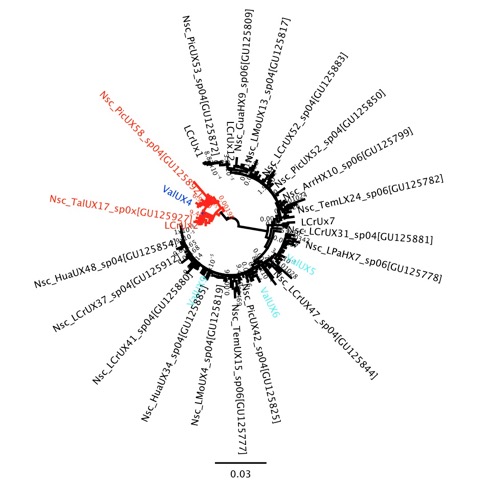
There is nothing too clear about the figure I’ve pasted in here, that is partly intentional because I’m still figuring some things out. But, this figure represents the first new step in understanding the genetic/species patterns along the coast of Chile that my lab has taken since our paper (Zakas et al. 2009) came out in MEPS. New lab technician Kelly Laughlin has been coming up to speed very quickly over the past 4 weeks, and the first sequence data she collected included about 24 individuals of Notochthamalus from near Valdivia, Chile. This is about 500km further south than our survey in the past, and I wondered if the genetic cline that we observed would go to fixation for the southern haplogroup. In the image above, the southern haplogroup is red, the northern haplogroup is black, and in blue (only a small number are shown because of the scale of the tree) are individuals from Valdivia (6 in the red clade, 12 in the black). What this tells me is that the relative B/A frequency in Valdivia is exactly the same (about 1/3) as at Pichilemu, Las Cruces, and Punta Talca far to the north.
This suggests that all of the focus can return to the area around 31°S latitude, where there are tremendous shifts in physical oceanography, ecology, biogeography, and population genetics. But it is also a puzzling scenario for our tiny barnacle. My interpretation is that we are dealing with the equivalent of two incipient species being formed from one, and it remains to be seen what micromicrohabitat differences they have, but the ‘southern’ one simply doesn’t find that suitable microhabitat to the north of 31°S very often. Our upcoming genomic scan will help point us toward the factors that may be maintaining or promoting this differentiation, similar to John Grahame’s exciting work on intertidal Littorinids.
Groveling
28/01/11 09:08
My Ph.D. advisor, Cliff Cunningham, always said I had an extraordinary knack for politely asking other people to do work for me. I think that was meant as a complement! What he was referring to was the difficulty in getting specimens of creatures, especially marine creatures, from around the world for studies of genetic variation. Sometimes it is absolutely essential that one do their own field work: to understand the community, to ensure the work is done right, to develop new questions. But sometimes, you know what you want and you know where you need it, and you know it would be an hour of work for somebody who lives right there and 5 days of work for you, and so you have to ask.
I’ve been fortunate to find people to help in so many such situations, and have been refining my spanish in the last few days to obtain some additional samples for my work in Chile, from a more remote location. Hopefully my conjugations make it clear that there is some urgency to these collections, since we would like to be sequencing them this spring. I’d love to travel to the border of Chile and Peru, but right now it makes sense to send a student out with a sharp knife and a vial full of ethanol, and pay them well so I can sleep a little better at night!
We have had some interesting results in the lab of late: in particular, the rotation project of Christine Ewers has opened up a new project that somebody can work on. A few years ago, we published sequence data from a range-wide survey of the minnow Notropis lutipinnis (yellowfin shiner), and showed that one mitochondrial gene (COI) and one nuclear gene (transferrin) told different stories about the historical ecology of this species. I assumed that the mtDNA was probably the safer bet, and that the nuclear gene was responding to differential environments among locations, specifically that the Flint and Altamaha drainages might have different microbial communities, or some other environmental component, than the Appalachian drainages in the rest of the study.
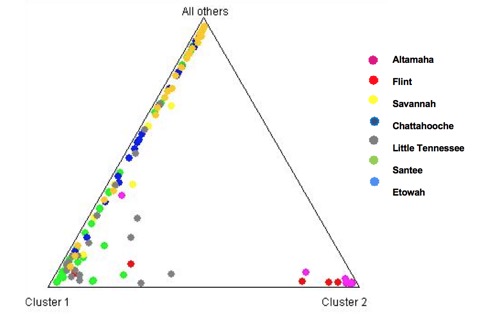
Christine came in and in less than 6 weeks managed to genotype all the individuals in that study (Scott-etal2009) with four microsatellite loci (we have more that will work, too). What these data showed pretty convincingly (see the Structure diagram above) is that the nuclear gene studied previously is not an outlier, because these additional data also suggest some historical demographic isolation of these more southern populations from those closer to the Tennessee and North Carolina and South Carolina borders. We’ll need to sample many more individuals to ensure this pattern is robust, but it looks like there are more mysteries to unlock in the biogeography of southeastern aquatic habitats.
I’ve been fortunate to find people to help in so many such situations, and have been refining my spanish in the last few days to obtain some additional samples for my work in Chile, from a more remote location. Hopefully my conjugations make it clear that there is some urgency to these collections, since we would like to be sequencing them this spring. I’d love to travel to the border of Chile and Peru, but right now it makes sense to send a student out with a sharp knife and a vial full of ethanol, and pay them well so I can sleep a little better at night!
We have had some interesting results in the lab of late: in particular, the rotation project of Christine Ewers has opened up a new project that somebody can work on. A few years ago, we published sequence data from a range-wide survey of the minnow Notropis lutipinnis (yellowfin shiner), and showed that one mitochondrial gene (COI) and one nuclear gene (transferrin) told different stories about the historical ecology of this species. I assumed that the mtDNA was probably the safer bet, and that the nuclear gene was responding to differential environments among locations, specifically that the Flint and Altamaha drainages might have different microbial communities, or some other environmental component, than the Appalachian drainages in the rest of the study.

Christine came in and in less than 6 weeks managed to genotype all the individuals in that study (Scott-etal2009) with four microsatellite loci (we have more that will work, too). What these data showed pretty convincingly (see the Structure diagram above) is that the nuclear gene studied previously is not an outlier, because these additional data also suggest some historical demographic isolation of these more southern populations from those closer to the Tennessee and North Carolina and South Carolina borders. We’ll need to sample many more individuals to ensure this pattern is robust, but it looks like there are more mysteries to unlock in the biogeography of southeastern aquatic habitats.
Climate and Weather
03/01/11 11:10
Given my last entry was about the weather, I wanted to be sure and point out that some of our funny weather is because of one of the short, fascinating cycles in the climate (something that climate skeptics often confuse is weather and climate). Seems we are going through a La Niña this winter, which should mean in general drier and warmer weather for Georgia. However, we also got a really nice snowfall late on December 25, so maybe we can have the best of both worlds for the next few months: good weather punctuated by beautiful snow!? I’ll keep my fingers crossed.
In the meantime, these oscillations in sea surface temperatures in the equatorial oceans keep me thinking of my barnacles down in Chile. I’ve just gotten word that the next shipment is about to arrive, just in time for new technician Kelly Laughlin to start in the lab. She will be working with others in the lab to get these specimens prepped for our Illumina next-generation sequencing efforts, hopefully to be done by March. My goal of March is set partly by what can be done by my colleagues down south, and partly by what I’m hoping people like student Christina Zakas can do before graduating, which is to help teach me all her tricks for SNP discovery and genome organization!
In the meantime, these oscillations in sea surface temperatures in the equatorial oceans keep me thinking of my barnacles down in Chile. I’ve just gotten word that the next shipment is about to arrive, just in time for new technician Kelly Laughlin to start in the lab. She will be working with others in the lab to get these specimens prepped for our Illumina next-generation sequencing efforts, hopefully to be done by March. My goal of March is set partly by what can be done by my colleagues down south, and partly by what I’m hoping people like student Christina Zakas can do before graduating, which is to help teach me all her tricks for SNP discovery and genome organization!