manuscript
Representation
19/02/14 12:04
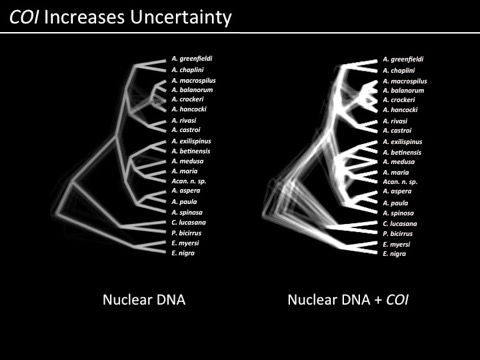
When I first started doing science, visualization of our data and results was a bit easier. Sequence a gene from 20-30 individuals, generate a phylogenetic hypothesis, make it look pretty, Figure 1. As we have moved to information from more and more loci (or greater numbers of samples, or any other factor of additional complexity), there have been ways developed to summarize the results visually for the purposes of proving to the audience of a journal article that you are on the right track in terms of interpretation. For example, former postdoc (and now faculty at Texas A&M - Galveston) Ron Eytan used these images to show which parts of a phylogeny were consistently supported and which were not (an image that overlays many reconstructions of a phylogeny from the same data):
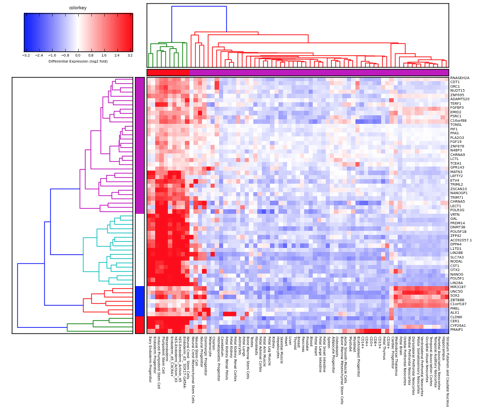
and we are now becoming familiar with things like transcriptome heat maps to illustrate over/under expression of particular genes:
and to an extent, we have just become accustomed to more complicated representations of data and results residing only as online supplemental files. In other words, often the most important components of science are no longer able to be represented in the standard peer-reviewed journal article. We are at this point often having to take into consideration representation of science through multiple filters: how will it communicate to the audience? what if that audience is looking only at a photocopy, or black-and-white print of my article? what if that audience is unable to afford the journal I submit to?
but what I’m grappling with today, is what if there is no single view, even if it is a figure with multiple panels, that can explain your results the same way that having an animation, in particular an interactive animation, could do?
the above is a representation of fitting in silico particles (representing barnacle larvae) to a coastal oceanographic system with environmental selection on the two lineages of barnacle that we are studying. The bottom plot shows frequencies of the northern and southern types plotted against latitude, and as the movie plays it shows how the oceanography and selection interact to fit simulated data to empirical data. This alone is a daunting amount of information. But we can vary so many parameters: depth of larval release, time in the water column, strength of selection, cost functions, the fit to over 100 distinct loci for which we have data, inter annual variation. There is almost no way to represent the complexity of this story - adequately! - in the illustrations we typically put together for a science publication. So it will probably end up that many of these movies will be online supplements. But I wonder: when will our expectations of “publishing” science catch up with the technology with which we DO science?

and we are now becoming familiar with things like transcriptome heat maps to illustrate over/under expression of particular genes:

and to an extent, we have just become accustomed to more complicated representations of data and results residing only as online supplemental files. In other words, often the most important components of science are no longer able to be represented in the standard peer-reviewed journal article. We are at this point often having to take into consideration representation of science through multiple filters: how will it communicate to the audience? what if that audience is looking only at a photocopy, or black-and-white print of my article? what if that audience is unable to afford the journal I submit to?
but what I’m grappling with today, is what if there is no single view, even if it is a figure with multiple panels, that can explain your results the same way that having an animation, in particular an interactive animation, could do?
the above is a representation of fitting in silico particles (representing barnacle larvae) to a coastal oceanographic system with environmental selection on the two lineages of barnacle that we are studying. The bottom plot shows frequencies of the northern and southern types plotted against latitude, and as the movie plays it shows how the oceanography and selection interact to fit simulated data to empirical data. This alone is a daunting amount of information. But we can vary so many parameters: depth of larval release, time in the water column, strength of selection, cost functions, the fit to over 100 distinct loci for which we have data, inter annual variation. There is almost no way to represent the complexity of this story - adequately! - in the illustrations we typically put together for a science publication. So it will probably end up that many of these movies will be online supplements. But I wonder: when will our expectations of “publishing” science catch up with the technology with which we DO science?
Baby It's Cold Outside
13/12/10 09:17
We woke in Athens to -6°C and winds, which made me really appreciate my building more than usual by the time I arrived after the 10 minute bike ride to get here. Figured it was time I updated before the semester is completely gone. Our lab did make it out to celebrate the end of a pretty good semester, we all (well, almost all, there was a final the next day...) headed to Trappeze to warm our insides and toast to our shared dislike of leafblowers. Most of all, it is good to have a lab full of people who get along and keep their heads, and maintain their enthusiasm in sweet and hard times about their passion for science. It was kind of a rough week in that regard, we had all together three papers rejected in recent days (but one accepted). The other papers will find the light soon enough.
Speaking of papers that need to find the light, I offer the following linked manuscript up to the science gods. This is a short paper that was orphaned from my dissertation, and this summer I challenged myself to write a manuscript on these data in one day. I actually took two, or parts of two, days knowing that it was only a tiny descriptive study of a limpet and perhaps no more interesting than the Genbank numbers contained within, but I guess I hope the data will be useful to somebody who is starting their graduate work and is considering work on Tectura testudinalis. David Reid is a wonderful colleague who I met at the CORONA meetings, and he was kind in his rejection, so I’m going to put this one into a web bottle and cast it to sea.
Wares_Tectura
The paper isn’t wrong, just insufficient. But the limpets are nice, so good luck little paper.
Our final news for the lab would be that we managed to earn three awards at the Genetics Department holiday party, I can’t remember which categories Christine’s entries were in but the vegan chickpea-spinach Indian-style dish I made got a blue ribbon in the vegetarian category, I’ll be sure to dogear that page in my cookbook!
Also if you are looking for entertaining reads on evolution and/or bees, check out the work of Jay Hosler (an image is on the “Invert Comics” page), a biologist who has combined his knowledge and skills as a cartoonist to generate some pretty interesting outreach.
Speaking of papers that need to find the light, I offer the following linked manuscript up to the science gods. This is a short paper that was orphaned from my dissertation, and this summer I challenged myself to write a manuscript on these data in one day. I actually took two, or parts of two, days knowing that it was only a tiny descriptive study of a limpet and perhaps no more interesting than the Genbank numbers contained within, but I guess I hope the data will be useful to somebody who is starting their graduate work and is considering work on Tectura testudinalis. David Reid is a wonderful colleague who I met at the CORONA meetings, and he was kind in his rejection, so I’m going to put this one into a web bottle and cast it to sea.
Wares_Tectura

The paper isn’t wrong, just insufficient. But the limpets are nice, so good luck little paper.
Our final news for the lab would be that we managed to earn three awards at the Genetics Department holiday party, I can’t remember which categories Christine’s entries were in but the vegan chickpea-spinach Indian-style dish I made got a blue ribbon in the vegetarian category, I’ll be sure to dogear that page in my cookbook!
Also if you are looking for entertaining reads on evolution and/or bees, check out the work of Jay Hosler (an image is on the “Invert Comics” page), a biologist who has combined his knowledge and skills as a cartoonist to generate some pretty interesting outreach.