Chthamalids
07/08/15 12:01
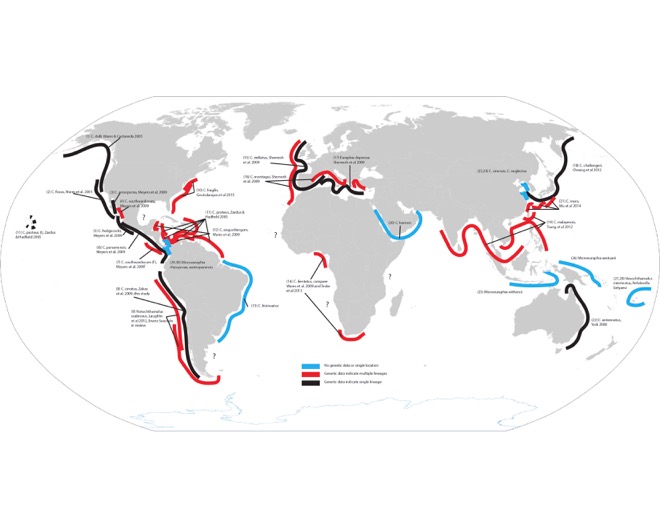
This tangled looking map is a laborious endpoint of me trying to put together what I know of diversity in the genus Chthamalus and close relatives (this includes some doubt about “close relatives” and whether Jehlius should really be Chthamalus as Darwin named it, whether the Euraphia and Microeuraphia are just clades within Chthamalus, and so on, and so it is still at best incomplete). I have a larger version of course if you want to see that. The black lines represent the studies where a nominal species has been evaluated with enough genetic data to confirm that it is, in fact, just one “thing”. The blue lines are taxa with insufficient information - maybe a single site, or none at all. And the red lines represent lineages within species that have been identified with genetic data, but have yet to be formally described as new species (and maybe that will never happen?). There are some interesting low-hanging fruit, depending on your access to certain parts of the world. For example, my work on C. dentatus that was published with Sabrina Pankey and Yair Achituv as co-authors contains very distinct diversity from the populations of C. dentatus in South Africa published by Teske et al. 2013. Benny Chan and colleagues have obviously identified a tremendous amount of diversity in southeastern Asia. Annette Govindarajan just found 3 cryptic lineages along the densely-studied east coast of North America! John Zardus showed us just how diverse “C. proteus” is in the Caribbean, and of course our work in the Chilean distribution of Notochthamalus is what had me thinking about this again.
I’d like to figure out how to make this build-able in R, so that as new data (and clades) are discovered the map can be auto-updated. For now that is slightly out of my reach in skill and time. But this is a single genus of barnacle that exhibits so much diversity genealogically, yet so little phenotypically. It is really just a phenomenon of calcareous beasts that distribute themselves everywhere, and diversify under mysterious processes. Diversity is a process, not a number. It is practically infinite in that regard (which makes it all the more stunning what a good job humanity is doing of messing up this process).
All your base are belong to us
15/08/11 10:00
It occurred to me that we are very close to having whole genomes for 48 microbial strains, and 50x coverage for a different set of 48 microbial samples from coral reefs in the Keys, and 50x coverage across each of 16 populations of RAD-tagged barnacles from Chile. By the end of the year. Something like 200,000,000,000 nucleotides that we have to analyze and make sense of in the coming year. If each one was a penny, stacked on top of each other it would just about reach to the moon. Good thing they aren’t that big.