Ups, Downs.
My plans were interrupted. First, by good news - my work on Pisaster is recommended for NSF funding along with colleagues Mike Dawson, Lauren Schiebelhut, Ian Hewson, and Pete Raimondi. Fantastic news that required some scrambling to address a few issues before anything could be finalized, and of course made trickier by me not being at my office. So, good news for sure and you'll hear more about that in the coming few years.
Then, the very next day (still in my first week at the labs), terrible news. One of the most incredible undergraduate students I've yet interacted with, Katelyn Chandler, passed away unexpectedly. She was only 20. I worked with her first as a teacher in my "Monsters" class, and then for 3 semesters in the lab where she learned how to do qPCR, genotyping sea stars, and eventually her Honors thesis on differential expression among EF1A genotypes of Pisaster. She was incredibly intelligent, engaging, diligent, and creative, and will be very much missed by all of us in the Wares lab.
Below is a photo I took of her presenting the poster she made with almost no help (in terms of design, content, and so on), discussing science that she had done much of the creative and intellectual work on.
Katelyn will receive a posthumous degree from the University of Georgia; in only 2 years at UGA she had already amassed tremendous number of credits, had completed her Honors thesis, had coauthored a scientific publication, and had touched the lives of very many.

Backlog Manifesto

small brazing project, JPW, 2011.
Summer is a time to gain perspective, at least in the academic world where we are fortunate to have a break from teaching and committee work (or at least, less of it). For many of us, this is a time for field work and travel to conferences, a time to catch up on the writing and analysis that was put off by the pressures of teaching. It's also when we make good on a lot of promises to kids, to family, to ourselves.
I just took my first real vacation in a long time. It was unrelated to work, unrelated to research. I did as little email-checking as I needed to, though there were a few tasks to keep juggling. And it is probably really trite to come back from vacation with a clarity that "life should be more like this", but there you have it.
I have been faculty for 10 years. I've literally lost count of how many PhDs, masters, and undergrad students I have shepherded through the process of research, how many different classes I have taught, even how many of my colleagues I was on committee to hire. I'm one of the semi-old guys now. I've had plenty of funding for my research, published a lot of it. I've been graduate coordinator for 4 years, and let me tell you that is both thankless and rewarding at the same time.
What concerns me about the academic life is the pressure to keep it up, to go more and more and more. Some of this is self-inflicted, of course, but we all know the numbers games that our university administrators are running, as well. For me, this has led to a lifestyle of juggling so many different projects that I recognize fully how each is being short-changed, and the people on them as well.
When I commit to a graduate committee, or to a student being in my lab, I should be committing sufficient time to that person that they will truly gain from the interaction – not that they will have to squeeze in a few half-baked moments with me when I spend half the time recuperating my understanding of what they are doing because it has been a few weeks since we discussed it and I've had to change mental focus so many times and haven't slept well and, and, and. Period. And I worry that isn't happening.
I'm not able to read much these days; I glean. I use close to a month of work hours to being grad coordinator, probably another month to basic administration of the lab (ordering, repairing, etc.) itself, and that is not at all dismissing the work my grads put in to the same tasks. I throw myself into teaching, and have been shuffled to new teaching assignments often enough that nothing is in balance (at least it isn’t getting stale!). And most importantly, I'm having a hard time finishing what I started because I feel the pressure to start something new. This is what I'm about to tell you, needs to stop.
So, though I am on about 3 pending proposals, and associated intellectually with a couple more, I am not submitting any more this year. Or maybe next. Funding is a precious commodity for research. My effort now is not scratching my head for ideas that might be fundable, to keep the treadmill at full speed, but to focus on the ongoing work and let it identify for me what ideas need to be funded, and how. What ideas arise organically, are useful, are critical to pursue? What are the priorities? Our planet, biodiversity, public health, as well as knowledge; each of us would come up with a different list of course.
I am helping 2 students finish their PhDs, and another 6 (unless I’ve forgotten somebody, which wouldn’t be surprising) that I'm currently on their graduate committees (in the past I have been on many more at once). That feels like enough, knowing all it takes in the last push. Including those who have recently finished - a masters student, postdocs, colleagues from other labs - I have at least 20, probably many more, manuscripts that are all in various stages of being written or generated. That is enough to keep me busy for years in and of itself! I want to do this science right, to do it the best possible way, rather than frantically pushing it away to get to the next thing. The people involved deserve my attention.
Notice how committee and commitment are related...?
This also means that travel is something that I am limiting. Travel is one of the true perks of a biologist, and I have excellent and dear colleagues all over the planet at this point. I get invited for talks, I get invited for intense one-on-ones about research or collaboration, and of course there are intriguing conferences where you get the opportunity to see more of the same people but in new! exciting! places where you still drink at the bar across the street from the conference center.... and I have to say no most of the time. If I have told you “no” recently, you aren’t alone. The environmental costs of flying are high, and this is a true priority to consider. The time (and financial) costs of travel are high as well, and I have to prioritize which ones are possible in the scope of all else I do professionally and personally. I think I would be interested to see how extended teleconferencing (Google hangouts, Skype, FaceTime, etc.) could replace some of these interactions – sure, you miss the local cuisine and some beers, and some research interactions are minimized – but we shouldn't neglect this magic that has transformed our professional lives in recent years, either.
This is a long justification for why you will hear "No" from me more often than in the past. I’m not doing anything other than recognizing my career as a long-distance run rather than a sprint, I’m trying to do right by those whom I am already committed to helping, I'm trying to do justice to the science I've already committed to, and I'm trying to do what is best for my family and personal needs as well. And I promise – and some of you know I’ve been up front about this in recent requests to come give a seminar, etc. – I won’t be offended if you tell me “No” as well.
In the Field
Hey, I love teaching. And there are parts of my job that I love. But most of those parts are concentrated in the few days a year when I’m working, but not in my building. Right now none of my projects are in active need of field sampling or monitoring or measuring anything, so I am living vicariously through my students. Christine led an expedition out to Sapelo last week, where the UGA Marine Institute put us up in a house on the marsh for 5 days of finding horseshoe crabs (for their Chelonibia epibionts - Christine’s dissertation beast), exploring the marsh, teaching my 8-year-old the taxonomy of fiddler crabs, a little beach time, and a lot of writing and excellent together time, all of it much needed.








It goes without saying I have about 200 more pictures as good as those or better. We got our quarry, maybe 1-2 more field trips for Christine on her way to doctorhood! We were all sad to leave Sapelo (the loss of a day in cleaning, driving, unpacking, exhaustion, back to building), but quickly got to look forward to the next trip - south central Georgia for a day on a river with Katie Bockrath and Mary Freeman’s fish crew, who helped get us a second round of mussel hosts collected on a beautiful day before a big storm. The timing is everything - water temperature, how high the stream is running, whether it is storming of course, whether we can round up a truck on short notice (not to mention a crew) - and this was almost effortless for once!






Thanks to everybody in the lab and otherwise who helped on these trips, in any way - these are the days I most feel like a biologist.
Seahawks

Almost on the same day, I had come across a series of TED talks about productivity in the workplace and how to best foster it. This is both for my own growth as a professional, trying not to get too caught up in the administrative tangle of a university while maintaining my trajectory as a biologist, as well as figuring out how best to work with students and colleagues.
One of the talks in particular was about leadership and productivity, and how engendering pleasant work environments - positive attitude, happy workers - leads to higher productivity, rather than the other way around. It triggered a lot of thoughts about how we use these ideas in graduate training, something that to me is a combination of medieval apprenticeship and modern-day productivity. We are training people how to do what we do, but we are also trying to get things done with them and through them. So how do we make this training experience best for students and mentors alike?
I try but I’m sure it could improve. I think I have happy students - at least, as happy as a grad student can be with all of the stresses of finishing a thesis and juggling the real life outside of these walls! But I know that as a department, we haven’t given much thought to how to work on the morale of the troops, how to calmly handle the fears and insecurities of generating ideas and backing those ideas up with data. This might be a good thing to think about before our next departmental retreat.
First Blog Stuff: A Green Lab
The unexamined conservation genetics lab is not worth leading. It's time to start thinking about the footprint of science, especially in labs that ostensibly are working toward conservation and an appreciation of the processes that generate and maintain biodiversity. I've brought my Kill-A-Watt into work to start evaluating ways to make my lab more efficient, with a smaller carbon footprint. I pay for carbon offsets, but that is just a financial trick to help me sleep at night (it doesn't work very well; I'm an insomniac!). This week I'm starting with the old chromatography refrigerator I inherited when I took over the lab. These giant glass-doored refrigerators can be useful, but we aren't running allozymes or SSCP gels in there, and really it may be that we don't need the thing at all. I've got it hooked into the Kill-A-Watt and will report on its cost (in dollars and in carbon molecules) soon. My goal is to reduce the kWh of my lab by 10% this month...I'm not sure if that is possible, so let's start crunching numbers. I'm working on the ballpark rate assumption of $0.10 per kWh, which may be a little high for Georgia but keeps the math easy (my friend Jason in Engineering Outreach tells me now that UGA's cost per kWh is $0.065).
Given that, after 24 hours that old chromo fridge used 5.49 kWh - about 1/100 of a typical monthly household energy use. That is actually surprisingly low, I think - that means it would probably cost about $197 a year to just leave it on. It isn't like there is an entire graduate fellowship lurking in that outlet. But, of course, add these things up across campus, or just across a lab...and that is a few pounds of CO2 (about 1.7 lbs CO2-equivalent per kWh). Now I'm going to check out my 'tip incubator', an old drying oven that we use just to make sure our pipet tips are dry - but that sits around heating the air ever so slightly, constantly. More on that tomorrow.
April 7, 2009
Okay, the incubator/oven set at lowest temperature point (holding at 29° in a lab that is probably about 24° ambient) - looks like that costs $28/year, about a sixth of the chromatography fridge (for a much smaller gizmo, that is also not really necessary for my lab). To establish some benchmarks for energy reduction, I guess I need to estimate kWh for a given time period, large enough to tell differences among the gizmos. The fridge used 5.49kWh in just over 24 hours, or about 2000kWh in a year. The oven on the other hand will use about 260kWh in a year (I didn't leave the K-a-W on for very long, so this is obviously ballpark).
A nice reference will be the one thing that I *know* I need for productivity from my lab - my computer. I have a Mac G5 with an LCD monitor, both about 4 years old. They are plugged into an uninterruptable power supply (UPS - fortunately, so I didn't have to shut anything down to plug in the K-a-W). We'll see just how much damage this setup does (to the environment, I mean - my eyes and wrists are certainly getting tired).
April 8, 2009
3.44kWh so far on my computer, in 16 hours. That is dismaying. For all intents and purposes it is almost equivalent to that giant refrigerator, despite having an LCD monitor that shuts off all night. But the computer doesn't - it is often running processes (number-crunching), or I access it from home, and for a few other reasons I leave it on for weeks at a time. There are certainly plenty of scheduling scripts that could be set up to shut it down each night, at least for a few hours, but I'm not sure that wouldn't mess with my (admittedly not high) productivity. I'm going to leave the K-a-W on for another day so it picks up a full day of me working on figures for the Notochthamalus paper, playing Guest Editor for Estuaries & Coasts, and of course checking si.com for Braves stories.
The trade-off may be that my lab currently has 8 computers - more than one per student. A couple are devoted to tasks, like the Gel Doc machine that is a real clunker. So I can't get rid of those, but that one in particular could be put on a shut-down schedule (except it is Windows, so that probably wouldn't work...). And another old machine is only serving information, that function could be transferred to another machine. Lots of options, for now I'll just keep crunching numbers.
Update: I found a python script that was able to rip the MySQL data from the old server G4 into HTML format. This is really dry reading for you, but I know someday at least *I* may find it useful. Anyway, the old G4 and its monitor are now shut down and unplugged from the wall, and I can access all that info (even with pictures) here on my main desktop. If you take nothing else away from reading this wiki-blog, its that learning scripting languages like perl and python is really, really useful in this information age. So, I'm going to assume that whatever kWh load (about 1980 kWh/year) my G5 puts on the world, I just saved by (hopefully permanently) unplugging my 9-year-old G4.
April 13, 2009
 The weather outside is gloomy, but through the fog - is that the Flying Spaghetti Monster? No, just a Monday looming. Final acceptance on Clare Scott's honors thesis came today, it is now "in press" at Ecology of Freshwater Fishes, good work Clare! This time of year seems to be manuscript-juggling: two dissertations, our paper at MEPS, a project starting up for J. Biogeog., and two Estuaries & Coasts manuscripts in revision. That means I need to be reminded every five minutes which project I'm working on.
The weather outside is gloomy, but through the fog - is that the Flying Spaghetti Monster? No, just a Monday looming. Final acceptance on Clare Scott's honors thesis came today, it is now "in press" at Ecology of Freshwater Fishes, good work Clare! This time of year seems to be manuscript-juggling: two dissertations, our paper at MEPS, a project starting up for J. Biogeog., and two Estuaries & Coasts manuscripts in revision. That means I need to be reminded every five minutes which project I'm working on.As for the Kill-A-Watt, the most recent info comes from my printer - which is a Lexmark that can print on both sides, so already a green step-up from our older laser printer. After 20 hours it had consumed 0.4 kWh, costing about $20/year to just leave it on. Certainly I could imagine figuring out a way to put it on a kill switch at night though, could cut that price in half. I'll add up those things later on in this project. The other thing that I measured over the weekend was my bank of MJ thermal cyclers. Obviously these are the bread-and-butter of the lab, so their usage is not really that negotiable. But, sadly, their job is to cycle back and forth between heating things and cooling things well past ambient, and they aren't insulated (they need to be able to change temperature rapidly, after all). So, 3 thermal cyclers under more-or-less normal usage for 94 hours used 8.32 kWh...about 775 kWh a year at a cost of about $77 (by my $0.10 guesstimate, which is a bit high - see post from a few days ago - but keeps math easy).
Now we are starting to get into the not-negotiable power consumption. For one, my ultracold freezer. That is necessary for long-term storage of valuable (hard to get again) DNA samples, and some enzymes. It churns pretty much constantly to keep the interior of the freezer at -80°C, and it is big - about 480L interior volume, and that is about 80% full (probably could be cleaned out and be 50% full). It runs 220V so I can't use the K-a-W, but online estimates from manufacturers suggest it is going to use 16.3 kWh per day, or about 6000kWh/year. Obviously these ultracolds are a big part of our footprint. The only way I can think of, off the bat, to deal with this is that there is a "departmental emergency" freezer in the same hall, that is almost empty and also running at -80°C. Maybe we need to negotiate a different arrangement? But, then, we've all had to deal with the crisis of a freezer that needs repair. At that time you need to move your irreplaceable samples immediately so they don't thaw, and so probably cannot wait for a freezer to get to temperature. At least, that is our fear talking...
SO the current tally: chromo fridge (now unplugged), incubator (now unplugged), G5, G4 (now unplugged), printer, ultracold, thermalcyclers used about 13000kWh/year. By unplugging the unnecessary stuff (for now), I'm helping...but there are a lot of gizmos to go. Assume the other 6 computers in the lab are also about 1800kWh/year - that is 10800kWh/year, a pretty massive load. So how do we cut back on computer load, without affecting computational power and analytical needs? That's the next thing to tackle. It will be, uh, a little easier once my two students finish up this summer...another G4 can be retired, for sure. But it is also important to figure out how some of these machines can be put on SHUTDOWN schedules, while still maintaining appropriate access to some data/servers at night.
Also, I sent this email today:
To whom it may concern,
every week I receive an issue of Columns, specifically addressed to me. I do find the information in Columns of value when I have time to read it, but I've noticed that dozens of copies in my building alone are immediately recycled or thrown away. Would it be possible for the University community to receive Columns as a formatted HTML email? Or to unsubscribe from the paper copy if they do not intend to read it? I notice in addition to the copies placed in each of our mailboxes (addressed to us, so our staff takes the time to sort them into mailboxes), there is a rack with extra copies in our mailroom.
At a time when the University is facing enormous financial burdens, changing the delivery default of Columns could be an enormous financial turnaround, not to mention an environmental shift that is in line with President Adams' stated goals. I'll look forward to your thoughts on this matter.
cheers John
There are lots of stones left to turn over at this University. If you are interested, check out http://gogreen.uga.edu/
April 16, 2009
Monitors are a big part of the power suckage of computers, but I'm surprised it isn't worse. I hooked the K-a-W up to the 24" LCD on our Mac Pro for a couple of days; looks like it is responsible for about 260kWh/year, or about $26 in operating costs (and about 445 lbs CO2e). So that leaves me pondering why the computer itself is so power-hungry at night? Well, it isn't set to go to sleep. So, whether I'm typing on it or not is immaterial. But I hesitate to set it to actually go to sleep, since that can grind analysis to a halt (this week for example I had a long PAML run and a long IMa run going; plus, I contact my G5 from home on occasion). There are ways to script a UNIX-based machine (like Mac OS) to check its processor load before going to sleep, but I'll need to do more research before applying this to my machine (for one, that link is 7 years old - OS X has changed a bit!). There are funny solutions to this problem, including the application Jiggler, which apparently mimics the mouse being jiggled often enough that the system won't go to sleep. This seems to be more for people who can't control the sleep/no sleep function, i.e. they aren't administrators. I had this problem with a Windoze machine while at UNM, I could *not* get it to stop going to sleep during a long analysis - so I put its mouse on a shaker table!
Anyway the trick for some of our machines may be to tell them explicitly to go to sleep - and warn folks not to use those machines for extensive analysis. The trick is, as haphazard as some of the development of my lab has been, it is hard to make major cuts without more control over the environment of the lab. Can't control the heat/air. The fume hood is OFF about 99.99% of the time because we don't use nasty chemicals - we just culture algae under a gro-lite in there. That's good because constantly-running fume hoods have to move so much air, they can be as energetically expensive as three U.S. households!
What I've cut so far - about 4500kWh/year - is about 7500 pounds of CO2 equivalent each year, and about $450. If you don't count heating and cooling but just the stuff we plug in, there is still another 12000 (computers), 6000 (ultracold), 6000 (other freezers and refrigerators), and probably another 6000 overall in other gizmos, or about 30,000kWh/year of energy use. So, for now I've met the 10% goal in a very coarse way. Many of us are able to ride our bikes in most days (I ride in at least 300 days a year)...more later...
April 20, 2009
As Earth Day approaches, continuing the energy-saving theme of this page seems good. At one faculty retreat a couple years ago, we began to discuss ways in which our department or building could reduce our footprint a bit. One thing that came up was a rant against using the elevator - at least, if you are reasonably healthy and not going too far or carrying too much, right? Well, I just came across this post that explains that elevators are one of your smaller concerns in a day's business. On to the next problem.
For now I'm leaving aside the consumption of kilowatt-hours by my lab; most remaining gizmos are somewhat necessary or infrequently used (and off/unplugged). The waste stream is probably the next good place to start. We try to maintain a very low-toxicity lab: no ethidium bromide (we use GelRed for staining gels), no radiation, extremely infrequent use of polyacrylamide or TEMED, and so on. Probably about 90% of our chemical usage is water, common salts, and naturally-derived enzymes in small volume. Our chemical usage could still be tweaked, but by and large mostly benign.
Plastics bugs me. Lots of virgin polypropylene. I estimated about a year ago, based on purchasing for the lab, that the common yellow tips used on 20µl and 200µl pipettors added up to about 4kg a year - the equivalent of 420 20-ounce soda bottles (at 28g each). Given the soda consumption of people in my lab (I'm just as guilty), I suspect we also drink and recycle about that many bottles a year. Which is worse? Washing and reusing the tips would definitely take a cut into our productivity (and would probably cause my students to mutiny), and recycling them is almost impossible (I think polypropylene is #5 plastic, which is not easily recyclable - and of course even if we think that our tips are only touching benign substances, would you buy a teething ring for your kid that advertised it was made of recycled scientific labware?). But, then, cutting out soda might put a notch in our productivity too, given the admitted caffeine addiction we've all got.
Wow, I didn't solve any problems with this information - bummer.
May 15, 2009
Wow, the end of a long semester and the beginning of summer. It is good to have a bit more leisure to consider how my projects are doing; just had my (almost) global phylogeny study on Chthamalus published in PLoS ONE, and today finishing up revisions on a Mark Vellend-inspired paper on genetic diversity in the Georgia Coastal Ecosystems domain.
I'm continuing the assault on electricity usage by my lab; right now our temperature-controlled room is not harboring any experiments or anything living (intentionally). Usually this 200-square-foot space is cooled to 18°C; it usually breaks about once a year because of the load of the first really hot days on the cooling system. If we aren't using the space, why not turn it off? That's gotta be worth something.
The question is, how does this energy usage affect the science? One could argue that my scientific program hasn't expanded to fill the resources I had available, or that I can only cut back in inverse proportion to my productivity. Only time will tell! We may have to devise a metric for productivity per kWh, perhaps the Hirsch index per kWh? Right now I'd be at about 0.00043 - no matter how it compares with others, that is a pretty unimpressive looking stat. I better get back to work.
Less is More

Everybody in the Genetics Department gets effectively the same 1000 sq ft lab to start with. They are functional but charmless - that is, they are functional if you are cloning and doing a lot of bench-top science. For bioinformatics, animal rearing, etc. the labs are less well planned, and if you actually want to talk to people in the lab without ducking under and around the shelving it gets awkward. So my renovations, maintaining all the potential for using the lab benches as they have traditionally been used but opening space up for our computational environment, have begun. We are capping all the water/gas/vacuum on this bench, having new outlets installed, and eliminating the sinks. Everybody is excited about it, and I think it will do a lot for the overall feel and ethos of the lab. We don’t do traditional bench-top genetics. I figure I’ll be in this lab for another 20 years, perhaps, so I may as well like it.
 (wrong picture somehow ended up here previously)
(wrong picture somehow ended up here previously)I am adding this update a couple weeks later because it is the same basic topic. I love that my lab is not merely/primarily a place of getting things done, but a place of learning and interaction. Inspired by what I’ve heard of the Santa Fe Institute, I encouraged my folks a few years back to use their windows to best advantage. We play music, we stop to talk about ideas, we have a constant supply of scrap paper to explain something to another colleague or student. That’s the way it is supposed to be. I can’t take any individual science project going on in my lab too seriously: they are barnacles, they are mussels, and so on. But the overall pursuit of understanding is a pretty beautiful thing some days.
Field Work
I just finished re-reading Steinbeck’s Log from the Sea of Cortez and two passages have to be shared.

“We knew that what we would see and record and construct would be warped, first, by the collective pressure....of our time and race, second ...by our personalities. But knowing this, we might not fall into too many holes - we might maintain some balance between our warp and the separate thing, the external reality. The oneness of these two might take its contribution from both. For example: the Mexican sierra has "XVII-15-IX" spines in the dorsal fin. These can easily be counted. But if the sierra strikes hard on the line so that our hands are burned, if the fish sounds and nearly escapes and finally comes in over the rail, his colors pulsing and his tail beating the air, a whole new relational externality has come into being - an entity which is more than the sum of the fish plus the fisherman. The only way to count the spines of the sierra unaffected by this second relational reality is to sit in a laboratory, open an evil-smelling jar, remove a stiff colorless fish from formalin solution, count the spines, and write the truth "D. XVII-15-IX." There you have recorded a reality which cannot be assailed - probably the least important reality concerning either the fish or yourself.”
and then...
“Our own interest lay in relationships of animal to animal. If one observes in this relational sense, it seems apparent that species are only commas in a sentence, that each species is at once the point and the base of a pyramid, that all life is relational to the point where an Einsteinian relativity seems to emerge. And then not only the meaning but the feeling about species grows misty. One merges into another, groups melt into ecological groups until the time when what we know as life meets and enters what we think of as non-life: barnacle and rock, rock and earth, earth and tree, tree and rain and air. And the units nestle into the whole and are inseparable from it. Then one can come back to the microscope and the tide pool and the aquarium. But the little animals are found to be changed, no longer set apart and alone. And it is a strange thing that most of the feeling we call religious, most of the mystical outcrying which is one of the most prized and used and desired reactions of our species, is really the understanding and the attempt to say that man is related to the whole thing, related inextricably to all reality, known and unknowable. This is a simple thing to say, but the profound feeling of it made a Jesus, a St. Augustine, a St. Francis, a Roger Bacon, a Charles Darwin, and an Einstein. Each of them in his own tempo and with his own voice discovered and reaffirmed with astonishment the knowledge that all things are one thing and that one thing is all things - plankton, a shimmering phosphorescence on the sea and the spinning planets and an expanding universe, all bound together by the elastic string of time. It is advisable to look from the tide pool to the stars and then back to the tide pool again.”
And so, we go to the field next month.
Wiki This Part Two

(the giant version is much better) and, reading more about the mountains I loved to hike and bike in when I lived there, I find out this astonishing fact: it is an uplifted coral reef, probably!
And so the amount we can learn when we are all communicating is pretty fantastic. My students in GENE 3000H (Honors Evolution) just finished their website on the interface between climate change and evolutionary processes, and it is really impressive to me how much they accomplished. This started a conversation among the folks in my lab about things like Wikipedia - traditionally, the bane of professors grading undergraduate research papers - being considered a component of scientific outreach. So my student Christine posts this page on turtle barnacles, starting from scratch. Katie was updating the page on Unionid mussels today. We are thinking this will be part of our weekly lab group, when we catch up with what each of us have been doing we’ll discuss the topics we have added to, and learned from. Obviously, Wikipedia is generally only the starting point for inquiry, but why not make it as complete as possible? Similar tools like the Encyclopedia of Life are also great things to contribute to (I haven’t tried this yet - next step!).
Budget Freeze
As it turns out, the University fired the few physical plant employees who were certified to work on these freezers last year. To save the school money, of course. Now, I pay the same 48.5% in overhead on my grants as always, but when my freezer dies the university doesn’t have my back anymore. A private contractor had to be called in, and long story short this will cost one of my grants nearly $6000. (Suggesting there is about $3000 in overhead that is covering the 10 burned-out 100W incandescent bulbs in the ceiling of my lecture hall? Or maybe that money could be used to repair the swimming pool on campus?).
Well, okay, I get it. State schools are strapped for money. Research costs money, and thankfully I have some NSF funds that will cover this so we can get back to work. But the math here doesn’t add up well.
In the meantime, my remaining freezers are packed to the gills and of course much of that needs to be re-organized, but we are also now at about the 8-year mark of my lab being open, and that means there is likely to be some stuff in there that just doesn’t need to be:

That’s one full 50-gallon trashcan just from one of the -20 freezers. There are some things that are hard to let go of, but need to be. For example, the box on top labeled ‘Tectura testudinalis’ are all of the samples (only about 35) that I collected of this limpet for my dissertation. But the sample is small enough that there is simply nothing useful to be done with them, beyond the freebie paper (see “Baby It’s Cold Outside”) I published on this blog last year!
Former and current students and postdocs: I assure you I’ve thrown nothing away that was properly labeled or appeared to be from the past, say, 4 years. But frostbitten 96-well plates with no labeling: gone!
Graduate Education
Good. Now put down the pipettor, and start learning to program. More and more, the work being done in labs like mine (and many of the evolution labs here in the Department of Genetics) involves data sets that are orders of magnitude larger than anything available when I got my Ph.D. Not only could you collect all of the data from my dissertation in one day of a well-organized sequencing effort, you would have to fill up the array with dozens of other dissertations as well to make it cost-efficient.... at a total cost of only a few thousand dollars.
All of that data means that it isn’t enough to have an interesting idea. You have to be able to manipulate large amounts of data, whether you are doing genomics or ecology or developmental biology, and this means you (prospective graduate student) need to become very familiar with languages or programming environments like R, python, perl. Twenty years ago you might be asked to be proficient in German or Spanish to get a Ph.D. Now it would be one of these languages, certainly. We spend more time in front of computers, and less time in the lab (fortunately, for a lab like mine, the amount of field work is about the same, though never as much as I’d like!).
Many fields are being transformed by computational approaches, and that is not a bad thing. It simply means more data. This is true in linguistics and literature as well as the sciences. So as you start to write your essays about what attracts you to our graduate program (I’m graduate coordinator, I expect to read about 40 of these essays in late December....), think hard about the ways you can learn to work with data quickly and efficiently. Make the data work for you, not the other way around. I say this from the rough experience of juggling a number of large data sets, each with their own quirks - now if only I could write a script that could write the papers for me as well....
Horseshoes

Another brilliant (but too short) trip to Sapelo. Christine Ewers is on another run of collecting horseshoe crabs for the Chelonibia (n.b. - as of June 2012, no page exists for this genus yet at Wikipedia, and barely noted in the Encyclopedia of Life) barnacles that grow on their backs. Horseshoe crabs come up to the beach at the highest tides to mate, so we were able to collect a few near sunset each day for her samples. Riley Wares helped out with the whirl-paks and enthusiasm!
For those of you who haven’t been to or worked at Sapelo Island, this is where UGA hosts its marine lab: certainly one of the more unusual marine labs I’ve been to. It has minimal infrastructure, but also minimal nearby development. Staff and researchers are housed in the buildings around R.J. Reynold’s old mansion, and we are well located for beach and salt marsh work (and play!). The island is separated from the coast by a 20-minute Department of Natural Resources ferry ride, a bargain at $1 per passenger.
Public
Fecundity 4

That’s Dr. Zakas next to me, successfully defended - in fact, she did such a good job on her dissertation I think the committee would have preferred to skip the defense and just head for the pub. Congratulations Christina!
Christina is going to be helping me in the coming months wrap up some analyses on the RAD-tagged data for Notochthamalus. I have to say right now I’m a little disappointed with the whole Nex-gen world, for the third time in as many attempts at getting 454 or Illumina data a huge portion of the data came back as junk (this time apparently a bubble in the line). I guess it will take a while for all the kinks to get worked out of these systems, and by then they’ll have invented something new!
Leaving!
John is heading on to a postdoc with Greg Moyer at the USFW facility in Warm Springs, GA.... as best as I understand this will be meta-analytical work on genetic patterns in the southeast, something that John - with his statistical and computational abilities in R and other environments - is very well suited to. Ron is going to continue working on our Serratia genomes while up at Yale working with Tom Near on big fish phylogenies - lots of fish, lots of genes, tons of data, again something ideal for his talents and interests. Good luck to both of you, and hope to see some manuscripts from you soon :)
All your base are belong to us
Gloves
Then the past 2-3 plates were bad again. Kelly and I did some sleuthing today and traced all the problems back to any sequencing that happened after April 27, 2011. On that day I sent a message out to the lab noticing that the autoclave gloves had gotten stuck in the seal at the top of the freezer (very close to where our pricey reagents live) and the freezer was thawing. Notice that even today the gloves are in the same place..... OK, I just moved them.

Anyway, I think our beloved BigDye must have sat at an elevated (thawing) temperature for 12 or more hours, and things haven’t been so good since. I hope it fixes things to buy a new $800 tube of reagent, because otherwise this constant problem-solving process is making my brain hurt.
A couple of links of interest: my colleague Sarah Gilman (of former Grosberg lab fame) has a nice paper out on the predictability of organisms shifting their range in response to climate change (other friends/colleagues like Leslie Rissler are in there too), here is a page pointing to that paper. And for anybody who likes to think about the response of the planet to past glaciation, the recent earthquakes in Maine are a nice reminder that nothing is static, it is always in flux.
Still Have Mojo
I love teaching the Evolution lab class at Georgia. I really do. But this class has led to more sleepless nights over the past 6 years than any other; why? Because maintaining a good flow of information, education, and fun in a lab class is tricky. And while failure is a large part of science, it doesn’t play well when you have a limited number of opportunities to teach and a number of topics you want to teach.
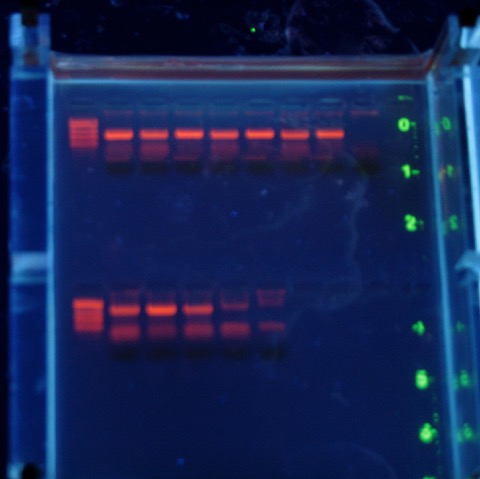
This year GENE 4230 will be repeating Peter Marko’s 2004 work on red snappers. This paper showed that much of what we buy at the store may actually be Lutjanus campechinus; but much of it is not. Fish fillets sold at red snapper prices are sometimes other (rarer) snapper species. Sometimes they aren’t snapper at all, but less valuable fishes. And all of that ambiguity leads questions about how best to manage this threatened stock. Currently, the fishery is closed on our Atlantic coast.
The problem? When I had Kelly double-check my protocol a couple of weeks ago to make sure the PCR would work, it did not. We tried a couple of different protocols, no dice. Finally I decided I needed to jump in and check some things. Long story short? Both our original primer stock, and the last batch of red snapper DNA we had used for this lab 2-3 years ago, were heavily degraded. They had been kept in the aging freezer in the teaching lab, so Genetics department head: there is something that we need to buy this year!
I’m pleased that I could quickly figure out what went wrong, and from the gel above it looks like the lab should work just fine (in case you are wondering, those are the short ‘universal’ fragment primers on top; the CB-12 and -13 primers from Marko’s paper below). There is a little smeariness in the negative control but I also didn’t change my tips in my rush to get this done today (Saturday....and it is 75° outside and sunny). No problem with the top set of reactions, that means I’ll be sleeping much better tonight.
Climate and Weather
In the meantime, these oscillations in sea surface temperatures in the equatorial oceans keep me thinking of my barnacles down in Chile. I’ve just gotten word that the next shipment is about to arrive, just in time for new technician Kelly Laughlin to start in the lab. She will be working with others in the lab to get these specimens prepped for our Illumina next-generation sequencing efforts, hopefully to be done by March. My goal of March is set partly by what can be done by my colleagues down south, and partly by what I’m hoping people like student Christina Zakas can do before graduating, which is to help teach me all her tricks for SNP discovery and genome organization!
Organization or the Lack Thereof
Today I’m coming to grips with the fact that a few hundred fairly important DNA isolations appear to have been lost somewhere in the last 3-4 years, they are some DNA samples from specimens collected in Chile in 2004. That is a bummer, because I may have an opportunity to get some next-gen sequencing done in the very very near future.... but I need some DNA! I have a technician starting in January to help organize these things, but it may be a busy next couple of weeks. Fortunately, I asked my students working on the first batch of Chile specimens to dissect away tissues and then keep the rest of the specimens in labeled vials. A real pain in the ass for them at the time, but a very much appreciated repository of information to me right now!
This has me thinking about how to organize samples in the future. Much of what we have is organized the way we would organize things when I was a grad student: it is in a lab notebook, or an Excel spreadsheet if we are lucky. That works fine for projects with samples in the low 100’s, but doesn’t work at all once the projects get larger, or have more loci, more locations, more times, and so on.
How complex do we want the process of curation to be? There are some really nice options, such as this LIMS plug-in for Geneious, which are worth considering as an all-in-one approach. However, the mental cost of getting started in that way is pretty high, both for me as advisor as well as for an incoming technician or graduate student. Most of the system only works well once you understand all steps of the process - and if you understand all steps of the process, you may also understand that Geneious doesn’t always handle each of these steps in the ideal way (though in general I’m a fan of that software).
For now I’m considering keeping things really basic - something like Excel or Numbers - which can then easily be uploaded into an actual database, either a baby one like Bento or a more complex one like that linked above. Either way it is clear it is time to get more savvy about what I’m freezing, because chances are good that any single freezer rack is containing a chapter for a graduate student, no reason for those hard-won samples to go to waste.
Technology
This week has been a good one in the Wares Lab, not least of which because a huge financial gamble has paid off, and is a great indicator of how population genetics and phylogeography is changing. I received some funding from the University of Georgia Research Foundation, intended for junior faculty to generate preliminary data for bigger fish (NSF, NIH) proposals. The entire goal of the award was to try out the Illumina Golden Gate genotyping technology with the Streblospio transcriptome that Christina Zakas is working with. We nervously sorted and re-checked the SNPs in the data, and weeded it out to a set of about 200 that we hoped would be applicable ... and more importantly, scorable. This species has been really recalcitrant when it comes to successful PCR. Fortunately, the Illumina technology doesn’t really go through PCR to get its genotype! In the end, 96 loci were chosen and the array was developed (again, at a big cost for a technology that we’ve never worked with or even seen worked with around here, on a species that we couldn’t even reliably amplify mitochondrial genes).
The data started rolling in this week, and it is phenomenal. Of course some of the loci don’t work well - but around 80% do, which is a better fraction than I’ve ever had when developing primers for nuclear loci in these non-model critters previously. And some DNA templates are not great quality, but close to 95% appear to be just fine. This puts us in position to suddenly be studying these tiny squishy mud worms at a global scale with 2 orders of magnitude more data than ever before. Money well spent, and we’ll be using the same technology on upcoming projects.
Meetings Meetings
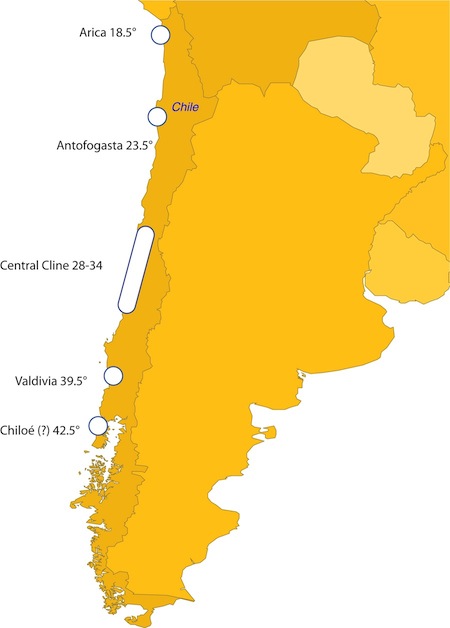
One trade-off involved with working out such details is that in a good collaboration, the workload is appropriately shared so that the best possible science is done most efficiently. In this case, my standing collaboration with Sergio Navarrete has led to a near-perfect situation where his postdoc happens to already be sampling many of the sites that we need samples from in the coming years. This is all going to work out well.... except now I have fewer good excuses for research jaunts to coastal Chile! Clearly, some of our face-to-face meetings will need to be in person, perhaps over a good plate of local seafood and some pisco sours.