biogeography
Chthamalids
07/08/15 12:01
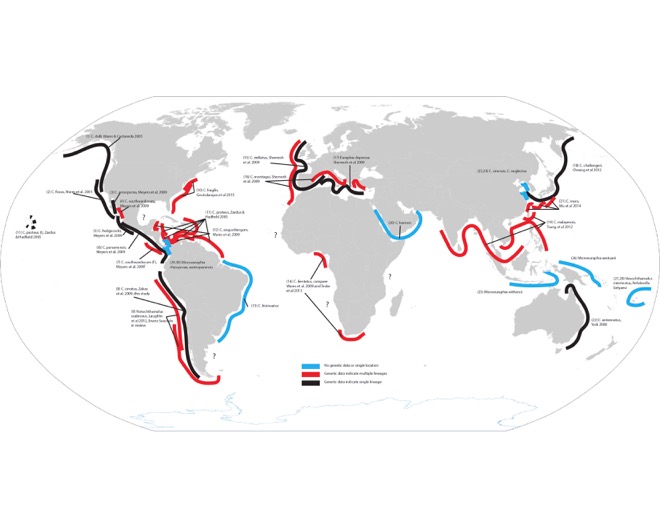
This tangled looking map is a laborious endpoint of me trying to put together what I know of diversity in the genus Chthamalus and close relatives (this includes some doubt about “close relatives” and whether Jehlius should really be Chthamalus as Darwin named it, whether the Euraphia and Microeuraphia are just clades within Chthamalus, and so on, and so it is still at best incomplete). I have a larger version of course if you want to see that. The black lines represent the studies where a nominal species has been evaluated with enough genetic data to confirm that it is, in fact, just one “thing”. The blue lines are taxa with insufficient information - maybe a single site, or none at all. And the red lines represent lineages within species that have been identified with genetic data, but have yet to be formally described as new species (and maybe that will never happen?). There are some interesting low-hanging fruit, depending on your access to certain parts of the world. For example, my work on C. dentatus that was published with Sabrina Pankey and Yair Achituv as co-authors contains very distinct diversity from the populations of C. dentatus in South Africa published by Teske et al. 2013. Benny Chan and colleagues have obviously identified a tremendous amount of diversity in southeastern Asia. Annette Govindarajan just found 3 cryptic lineages along the densely-studied east coast of North America! John Zardus showed us just how diverse “C. proteus” is in the Caribbean, and of course our work in the Chilean distribution of Notochthamalus is what had me thinking about this again.
I’d like to figure out how to make this build-able in R, so that as new data (and clades) are discovered the map can be auto-updated. For now that is slightly out of my reach in skill and time. But this is a single genus of barnacle that exhibits so much diversity genealogically, yet so little phenotypically. It is really just a phenomenon of calcareous beasts that distribute themselves everywhere, and diversify under mysterious processes. Diversity is a process, not a number. It is practically infinite in that regard (which makes it all the more stunning what a good job humanity is doing of messing up this process).
Quammen
27/05/13 12:37
If you are even a little bit interested in the travails of 19th-century naturalists and want to know more about the field of biogeography and what it can say about the current extinction crisis our planet is in, I highly recommend David Quammen’s The Song of the Dodo. A phenomenal book on diversity, conservation, biogeography, and evolution. The experience and wisdom involved in writing this book is enviable.
North Atlantic
07/10/10 10:46
It’s funny, I made my start as a population geneticist championing the cause of the North Atlantic Ocean, and I don’t think about it as often these days. Our careers creep in many funny directions, sometimes pulled by funding opportunities and sometimes by fortuitous collaboration.

The above image was snared from the website of Geoff Trussell’s lab at Northeastern University. Geoff is one of my many great colleagues that I know from interactions at the CORONA meetings and every year that I can make it to the Benthic meetings on the east coast (and really, who would want to miss these meetings? Nice one, Jeremy...). Anyway, the image is primarily of the dogwhelk, Nucella lapillus. A really beautiful snail, lots of color variation, interesting genetic patterns, part of my dissertation.
The work is finally being updated, with modern statistical (ABC) approaches, by my good friend and colleague Mike Hickerson and his student and postdoc (particularly lead author Katriina Ilves). The results are clearly starting to change. There are taxonomic controls on who survived glaciation and who didn’t - on the east coast of North America, at least - rather than larval life history controls, if the statistics tell the whole story. What is amazing is how big of an impact that paper had in 2001, and I suspect this paper will have a big impact in 2010.... and yet we still clearly know so little. Most of these species have still only been assayed for mitochondrial variation. The time will come soon when we can apply for funding to tackle the same community using variation across the entire genome.

The above image was snared from the website of Geoff Trussell’s lab at Northeastern University. Geoff is one of my many great colleagues that I know from interactions at the CORONA meetings and every year that I can make it to the Benthic meetings on the east coast (and really, who would want to miss these meetings? Nice one, Jeremy...). Anyway, the image is primarily of the dogwhelk, Nucella lapillus. A really beautiful snail, lots of color variation, interesting genetic patterns, part of my dissertation.
The work is finally being updated, with modern statistical (ABC) approaches, by my good friend and colleague Mike Hickerson and his student and postdoc (particularly lead author Katriina Ilves). The results are clearly starting to change. There are taxonomic controls on who survived glaciation and who didn’t - on the east coast of North America, at least - rather than larval life history controls, if the statistics tell the whole story. What is amazing is how big of an impact that paper had in 2001, and I suspect this paper will have a big impact in 2010.... and yet we still clearly know so little. Most of these species have still only been assayed for mitochondrial variation. The time will come soon when we can apply for funding to tackle the same community using variation across the entire genome.
Back in the Saddle
05/04/10 11:16
Well, if I thought things were busy in my last post - nearly 2 months ago - it seems somebody has stepped on the accelerator in the meantime. My office Mac had the new system installed, so that has taken me a while to find this document again and have something to report. But in the last 2 months, we’ve been pleased to get acceptances from 11 incoming graduate students, hooray! And I’ll be working with them again in our Intro Methods and Logic class this fall, so it will be good to get to know everybody.
Also, it seems I’m becoming more permanent around here, as my collaborations have become official: the Bio-Oce proposal with Jeb Byers and Jamie Pringle is funded and starting in June, and my collaboration with Jim Porter and Andrew Park (Ecology), Erin Lipp (EHS), and Katie Sutherland (Rollins College) was also just tabbed for funding on April 1 (we think it isn’t an April Fool’s joke....)!
So, for all the high-tech wizardry I’ve proposed in the past, ultimately I’m thrilled to be funded essentially to do biogeography and community ecology. That is what got me started down the academic path in the first place, really. Stay tuned!
Also, it seems I’m becoming more permanent around here, as my collaborations have become official: the Bio-Oce proposal with Jeb Byers and Jamie Pringle is funded and starting in June, and my collaboration with Jim Porter and Andrew Park (Ecology), Erin Lipp (EHS), and Katie Sutherland (Rollins College) was also just tabbed for funding on April 1 (we think it isn’t an April Fool’s joke....)!
So, for all the high-tech wizardry I’ve proposed in the past, ultimately I’m thrilled to be funded essentially to do biogeography and community ecology. That is what got me started down the academic path in the first place, really. Stay tuned!