Groveling
28/01/11 09:08
My Ph.D. advisor, Cliff Cunningham, always said I had an extraordinary knack for politely asking other people to do work for me. I think that was meant as a complement! What he was referring to was the difficulty in getting specimens of creatures, especially marine creatures, from around the world for studies of genetic variation. Sometimes it is absolutely essential that one do their own field work: to understand the community, to ensure the work is done right, to develop new questions. But sometimes, you know what you want and you know where you need it, and you know it would be an hour of work for somebody who lives right there and 5 days of work for you, and so you have to ask.
I’ve been fortunate to find people to help in so many such situations, and have been refining my spanish in the last few days to obtain some additional samples for my work in Chile, from a more remote location. Hopefully my conjugations make it clear that there is some urgency to these collections, since we would like to be sequencing them this spring. I’d love to travel to the border of Chile and Peru, but right now it makes sense to send a student out with a sharp knife and a vial full of ethanol, and pay them well so I can sleep a little better at night!
We have had some interesting results in the lab of late: in particular, the rotation project of Christine Ewers has opened up a new project that somebody can work on. A few years ago, we published sequence data from a range-wide survey of the minnow Notropis lutipinnis (yellowfin shiner), and showed that one mitochondrial gene (COI) and one nuclear gene (transferrin) told different stories about the historical ecology of this species. I assumed that the mtDNA was probably the safer bet, and that the nuclear gene was responding to differential environments among locations, specifically that the Flint and Altamaha drainages might have different microbial communities, or some other environmental component, than the Appalachian drainages in the rest of the study.
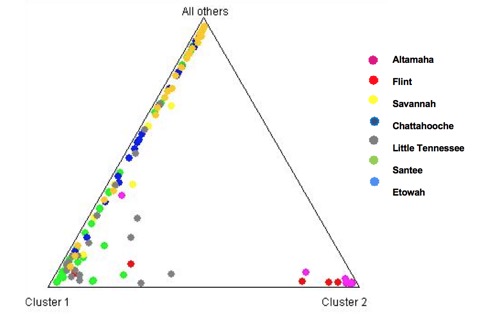
Christine came in and in less than 6 weeks managed to genotype all the individuals in that study (Scott-etal2009) with four microsatellite loci (we have more that will work, too). What these data showed pretty convincingly (see the Structure diagram above) is that the nuclear gene studied previously is not an outlier, because these additional data also suggest some historical demographic isolation of these more southern populations from those closer to the Tennessee and North Carolina and South Carolina borders. We’ll need to sample many more individuals to ensure this pattern is robust, but it looks like there are more mysteries to unlock in the biogeography of southeastern aquatic habitats.
I’ve been fortunate to find people to help in so many such situations, and have been refining my spanish in the last few days to obtain some additional samples for my work in Chile, from a more remote location. Hopefully my conjugations make it clear that there is some urgency to these collections, since we would like to be sequencing them this spring. I’d love to travel to the border of Chile and Peru, but right now it makes sense to send a student out with a sharp knife and a vial full of ethanol, and pay them well so I can sleep a little better at night!
We have had some interesting results in the lab of late: in particular, the rotation project of Christine Ewers has opened up a new project that somebody can work on. A few years ago, we published sequence data from a range-wide survey of the minnow Notropis lutipinnis (yellowfin shiner), and showed that one mitochondrial gene (COI) and one nuclear gene (transferrin) told different stories about the historical ecology of this species. I assumed that the mtDNA was probably the safer bet, and that the nuclear gene was responding to differential environments among locations, specifically that the Flint and Altamaha drainages might have different microbial communities, or some other environmental component, than the Appalachian drainages in the rest of the study.

Christine came in and in less than 6 weeks managed to genotype all the individuals in that study (Scott-etal2009) with four microsatellite loci (we have more that will work, too). What these data showed pretty convincingly (see the Structure diagram above) is that the nuclear gene studied previously is not an outlier, because these additional data also suggest some historical demographic isolation of these more southern populations from those closer to the Tennessee and North Carolina and South Carolina borders. We’ll need to sample many more individuals to ensure this pattern is robust, but it looks like there are more mysteries to unlock in the biogeography of southeastern aquatic habitats.